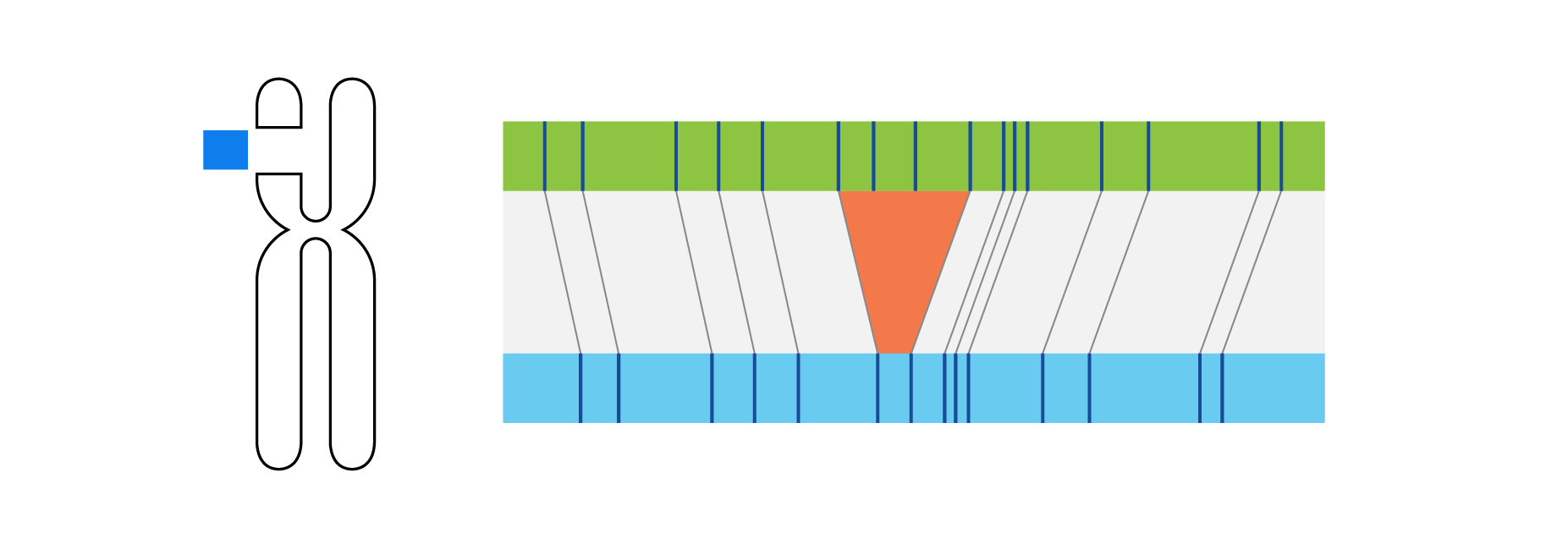
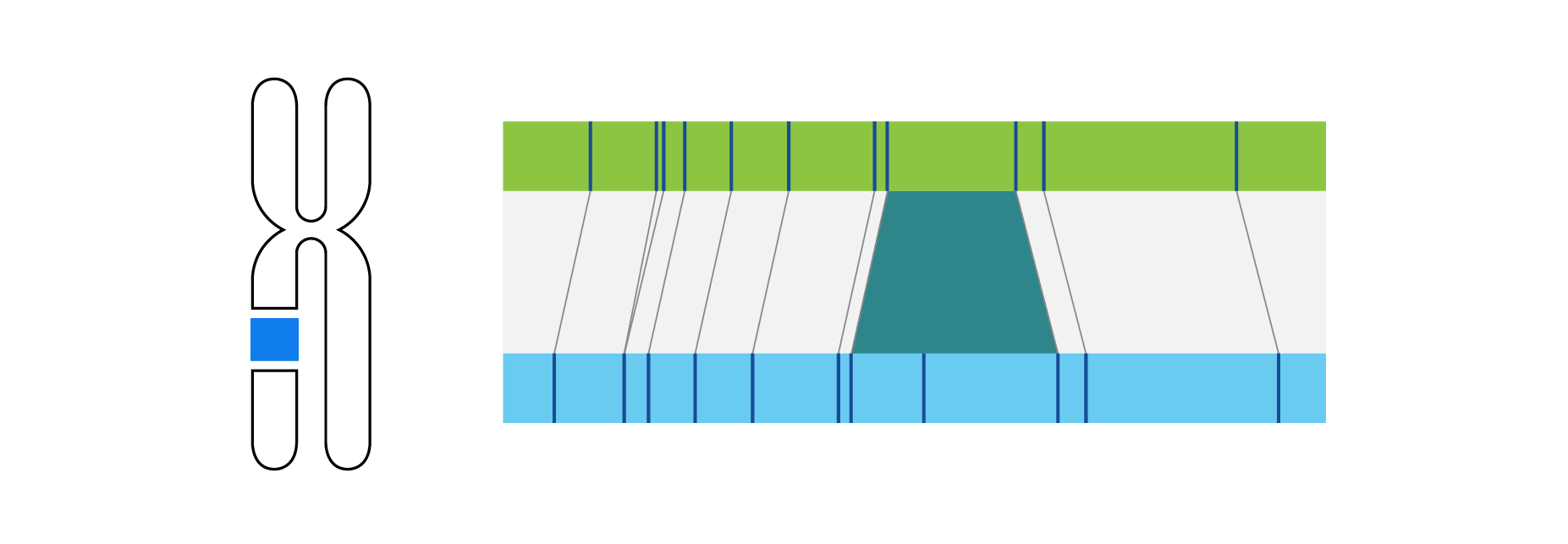
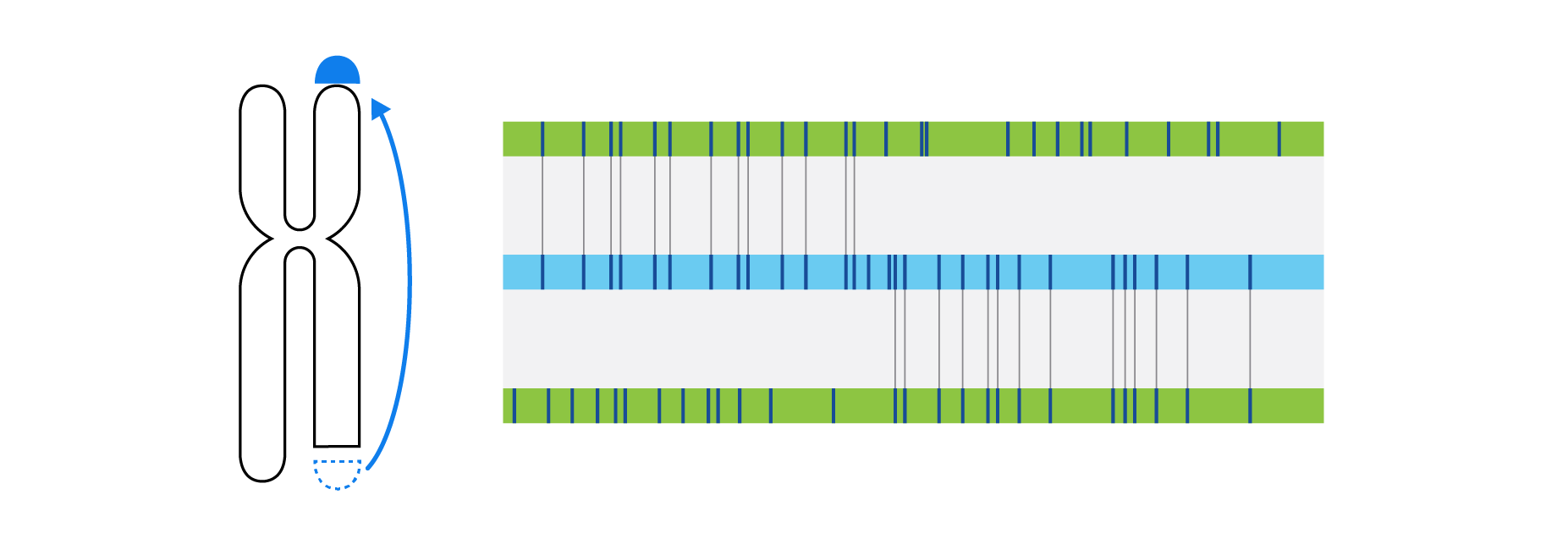
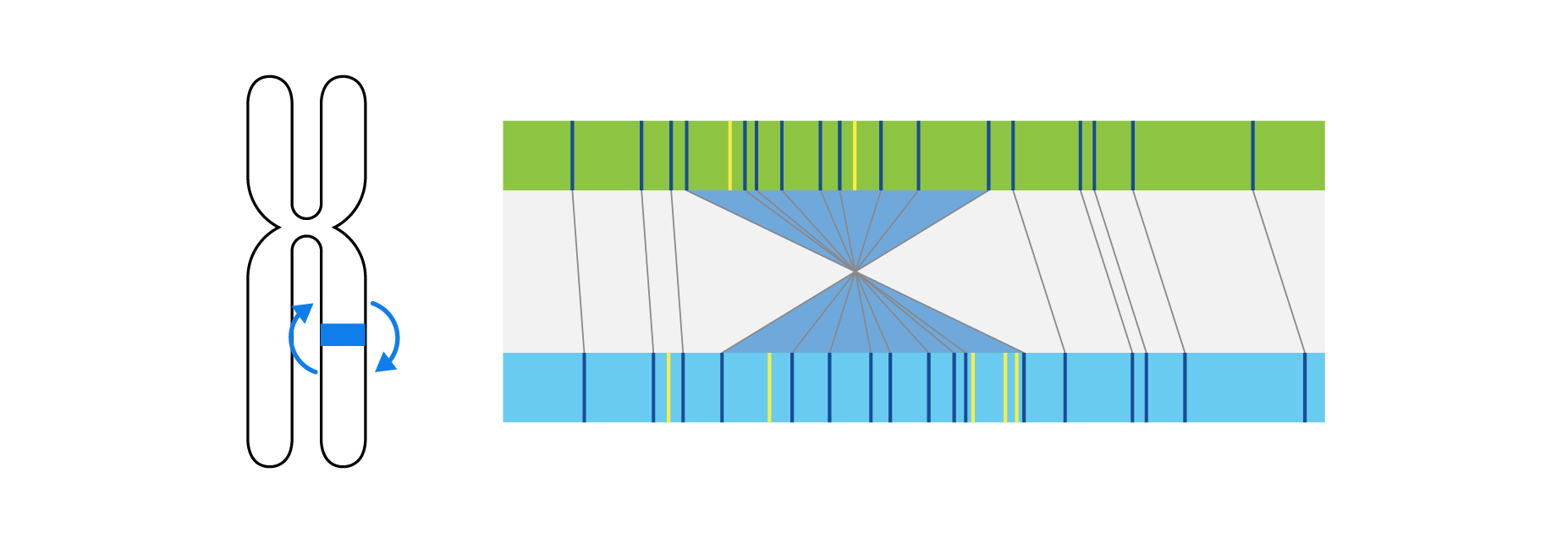
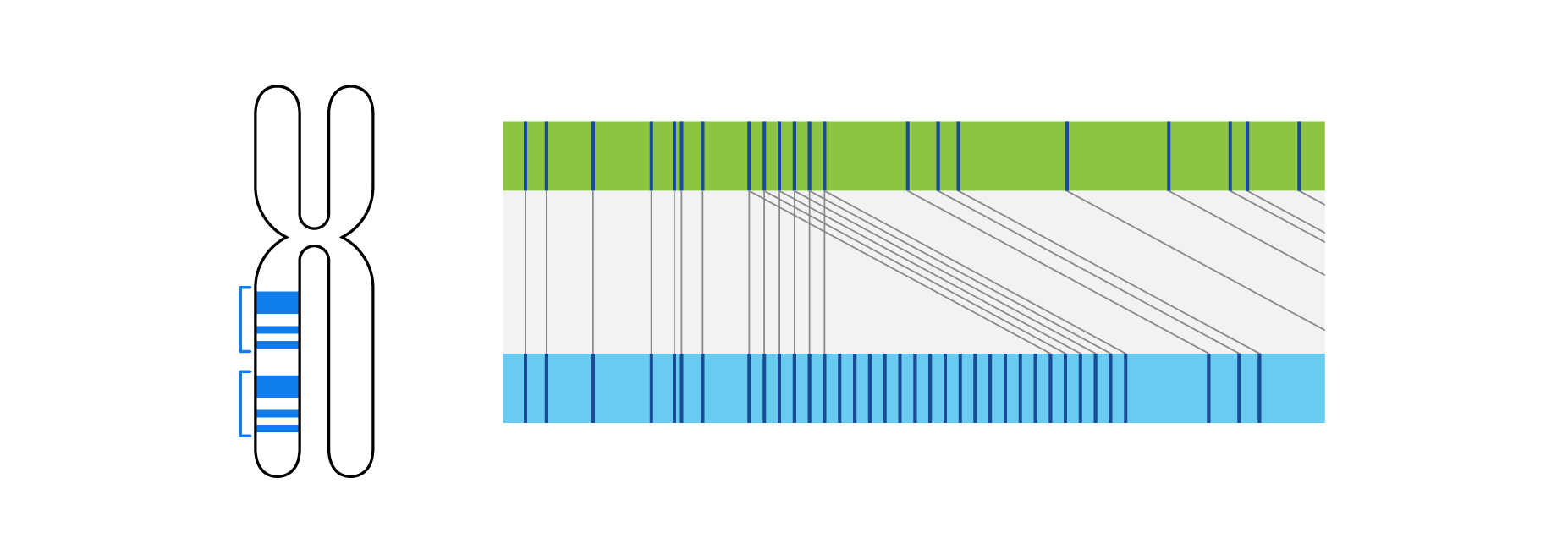
What is Structural Variation?
Structural variations (SVs), including chromosomal aberrations, are changes to the structure or number of chromosomes. Chromosomes are long DNA molecules containing the genetic material of living organisms. There can be extra or missing chromosomes, or chromosomes that have different types of rearrangements resulting in gains and losses of DNA, copy number changes, and balanced shifts in position. Any of these SVs can disrupt the normal function of a gene and contribute to the development of a broad range of genetic diseases and cancer. Unlike single nucleotide variants (SNVs) or small deletions and duplications (indels), SVs can range in size from a few hundred base pairs to thousands or millions of base pairs of DNA. Traditionally, SVs have been identified by karyotyping, fluorescence in situ hybridization (FISH), and chromosomal microarray (CMA).
Optical genome mapping (OGM) with Bionano genome mapping systems can identify all classes of structural variation by observing changes in label spacing and comparisons of order, position, and orientation of label patterns. Tab through the image carousel below to see how each type of SV is visualized with OGM.

The Role of SVs in Human Disease
SVs are a key part of genomic variation that shapes evolution.1,2 While SNVs contribute to 0.1% variation between humans, SVs contribute 1.5%.3 People can carry many structural variants in their genomes, where DNA segments are duplicated, deleted, inverted, or changed. In some cases, there are no detrimental health effects. However, these genomic structural variants are increasingly associated with disease: 4-9
- 25% of protein truncating mutations are a result of SVs
- SVs are 3x more likely to associate with a genome-wide association study signal than single nucleotide variants (SNVs)
- Larger SVs (>20 kb) are up to 50-fold more likely to affect the expression of a gene compared to SNVs
Explore the role of SVs in different disease-focused applications in more detail on the linked pages below.

Related Applications
Cytogenomics
Molecular Genomics
Genetic Disease
Oncology
Cell Bioprocessing
Learn more about OGM
Read a step-by-step summary of how OGM works.
Learn MoreFind the latest research in our Publications Library.
Learn MoreLearn from a specialist: Request a live overview of how OGM works and discuss how your research can benefit.
Request an OGM overview- Perry GH, Yang F, Marques-Bonet T, et al. Copy number variation and evolution in humans and chimpanzees. Genome Res. 2008;18(11):1698-1710. doi:10.1101/gr.082016.108
- Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 2013;14(2):125-138. doi:10.1038/nrg3373
- Pang AW, MacDonald JR, Pinto D, et al. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 2010;11(5):R52. doi:10.1186/gb-2010-11-5-r52
- Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445-449. doi:10.1126/science.1138659
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899-905. doi:10.1038/nature08822
- Talkowski ME, Rosenfeld JA, Blumenthal I, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149(3):525-537. doi:10.1016/j.cell.2012.03.028
- Collins RL, Brand H, Karczewski KJ, et al. A structural variation reference for medical and population genetics [published correction appears in Nature. 2021 Feb;590(7846):E55]. Nature. 2020;581(7809):444-451. doi:10.1038/s41586-020-2287-8
- Chiang C, Scott AJ, Davis JR, et al. The impact of structural variation on human gene expression. Nat Genet. 2017;49(5):692-699. doi:10.1038/ng.3834
- Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747-753. doi:10.1038/nature08494