Molecular Genomics
Unveil comprehensive structural variant profiles of cancer and genetic disease samples, and maximize pathogenic findings, with optical genome mapping (OGM).
Detection of structural variants (SVs) is essential in clinical and translational research. However, next-generation sequencing (NGS) methods fail to detect all classes and sizes of SVs, missing a significant amount of information critical to understanding disease biology.1-4
Studies have shown that short-read and long-read sequencing technologies miss a significant portion of SVs detected by OGM.2,3 Combine NGS with OGM to reveal a larger spectrum of genetic variants, generate a more complete profile of cancer and constitutional samples, and discover new actionable biomarkers.




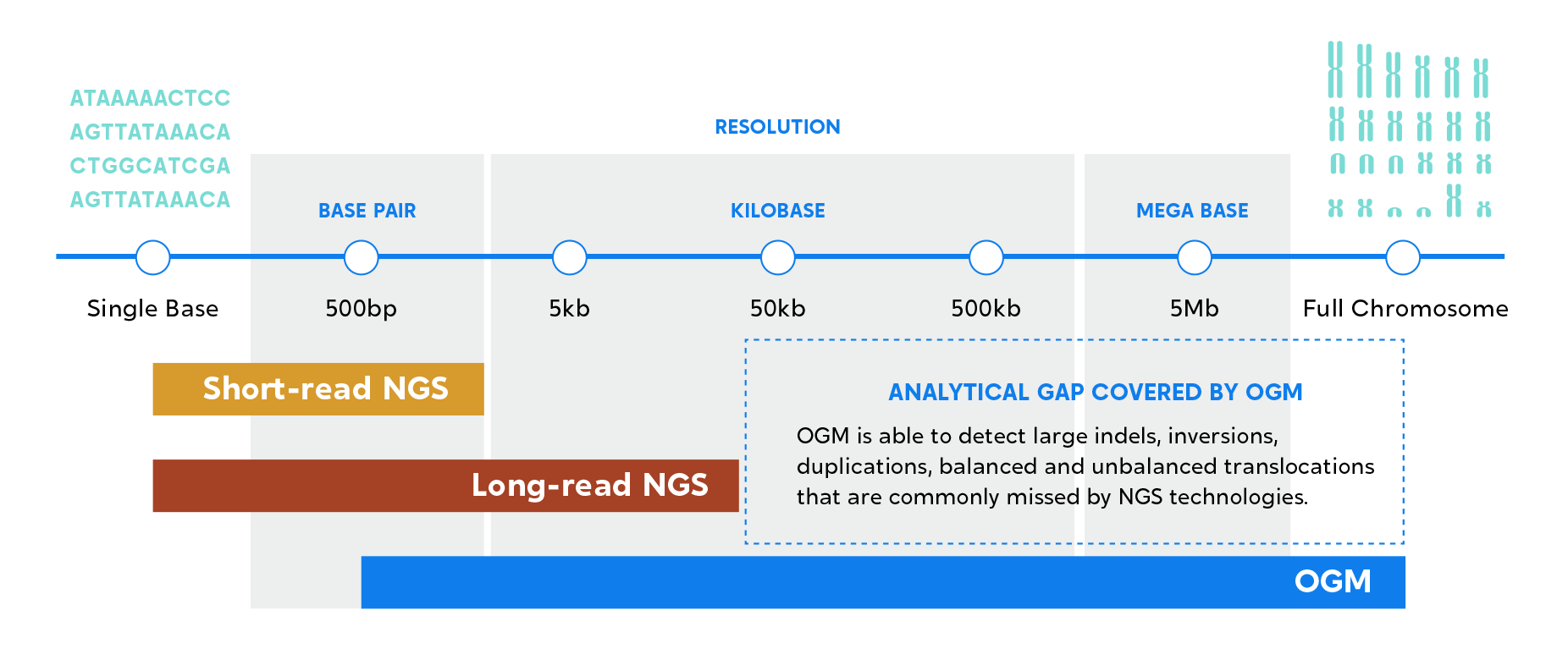
Combining OGM and NGS for resolution that ranges from single base pairs to chromosome scale enables the most comprehensive variant detection and view of the genome, suitable for novel biomarker discovery and translational/clinical research applications.
Combine NGS and OGM to achieve a comprehensive view of the genome, across all variant types and sizes.
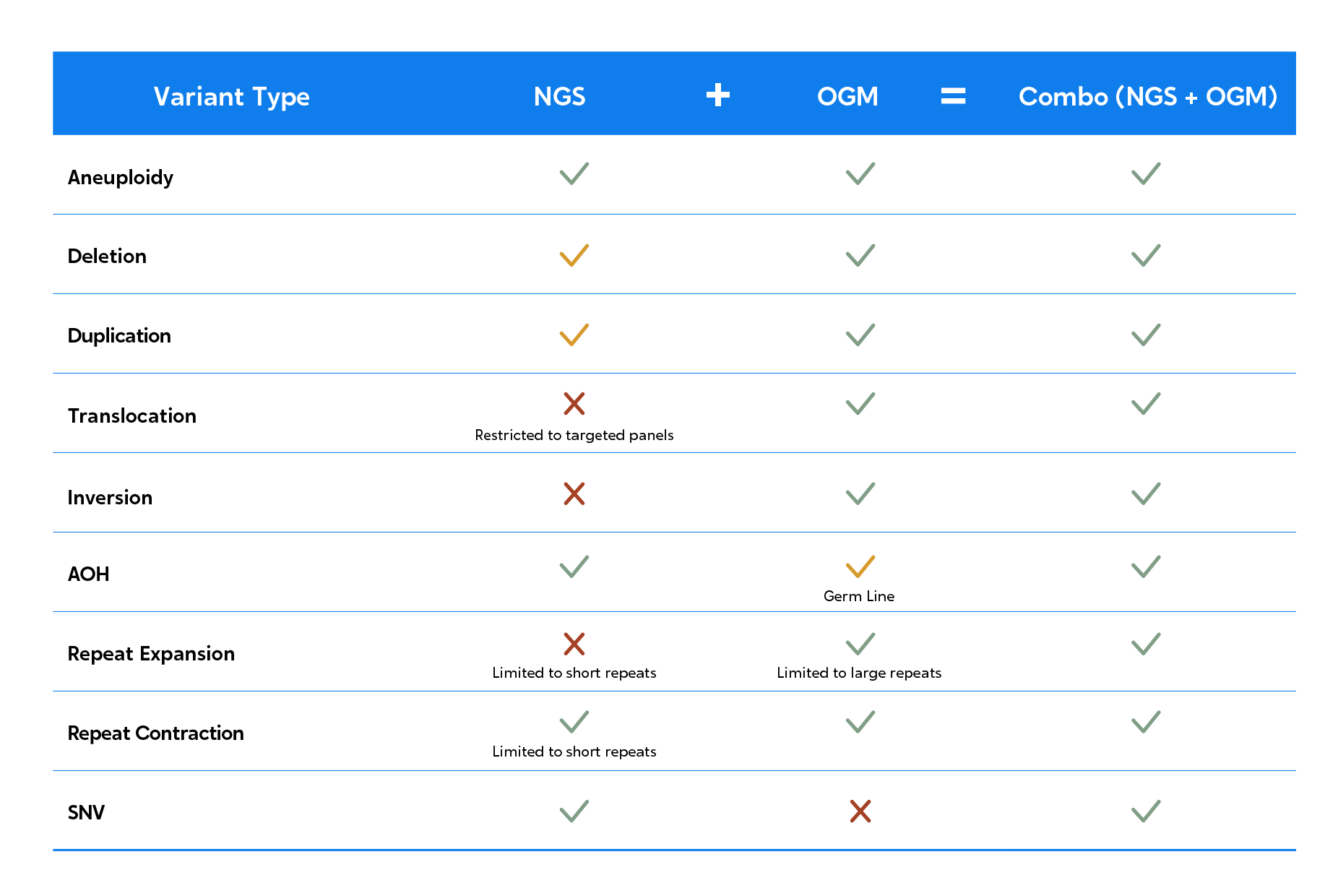
Combine the strengths of OGM and NGS to unveil a truly comprehensive genomic profile.
As this Nature study, from the Human Genome Structural Variation Consortium (HGSVC) demonstrates, OGM can reveal many SVs that are missed by NGS methods.1 Add OGM to your NGS data to achieve comprehensive genomic profile of samples, and maximize pathogenic findings
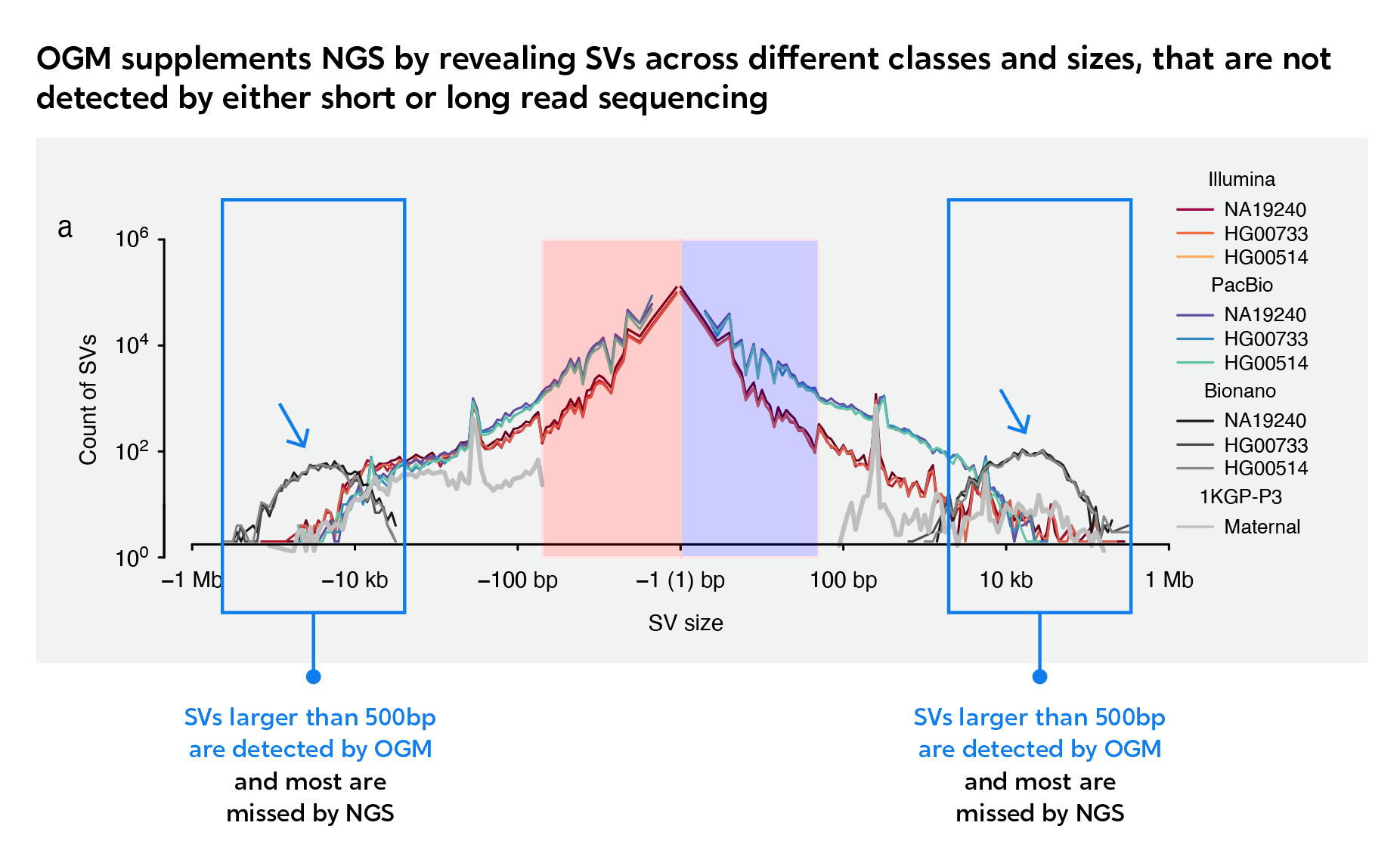
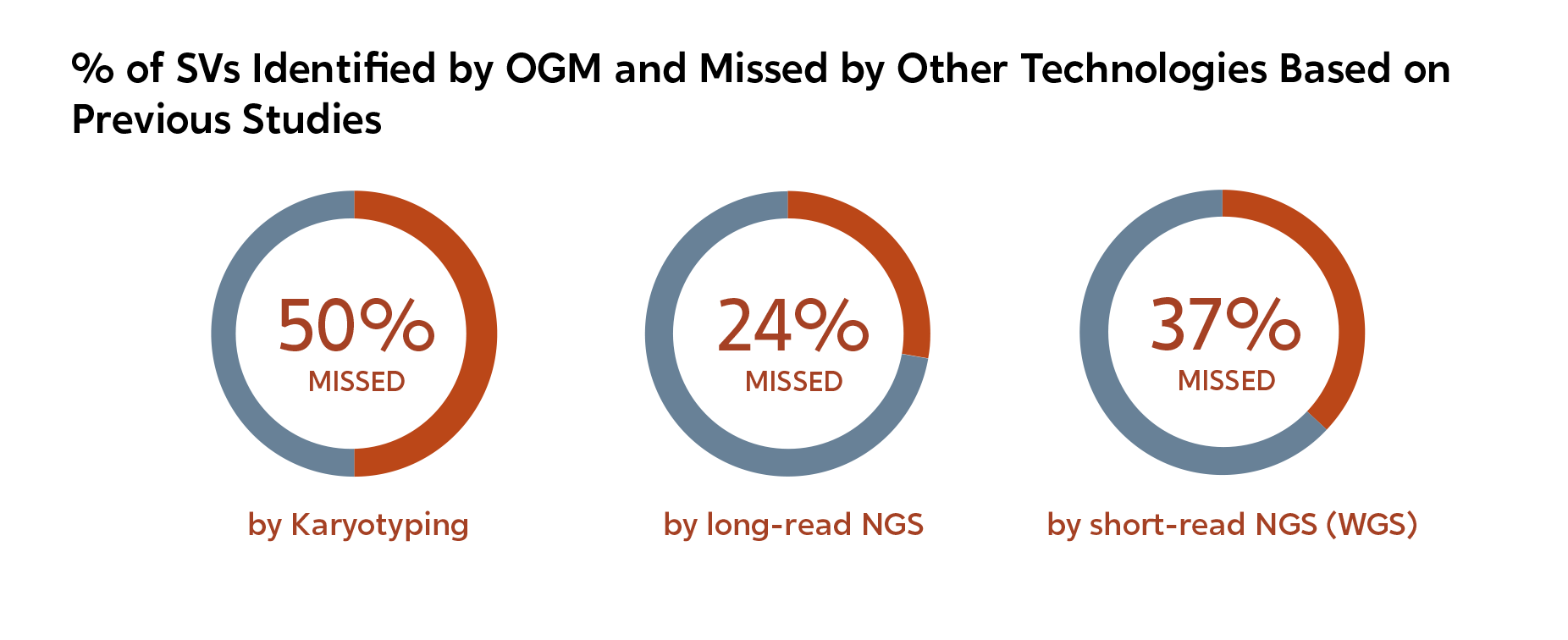
There are dozens of publications reporting SV findings from OGM that are missed by classical cytogenetics and NGS approaches. Here are a few quantitative examples:
Combine OGM and NGS to maximize variant detection across the full variant spectrum.
Complimentarity of Optical Genome Mapping and Next Generation Sequencing Panel (523 Genes) for the Comprehensive Evaluation of Myeloid Neoplasms
Watch videoOptical Genome Mapping: Hidden SVs in rare diseases
Watch videoWatch several scientific experts discuss the value of combining NGS and OGM across oncology and constitutional applications
Watch videosRead about what structural variations are and why they matter.
Learn MoreSee how OGM reveals structural variation in a way that has never been done before.
Learn MoreFind the latest research in our Publications Library.
Learn More
| Title | Source | Authors |
|---|---|---|
| Optical genome mapping unveils hidden structural variants in neurodevelopmental disorders |
Nature May 16, 2024 |
Isabelle Schrauwen, Yasmin Rajendran, Anushree Acharya, et al. |
| A comprehensive approach to evaluate genetic abnormalities in multiple myeloma using optical genome mapping |
Blood Cancer Journal May 3, 2024 |
Ying S. Zou, Melanie Klausner, Jen Ghabrial, et al. |
| Generation of three isogenic, gene-edited iPSC lines carrying the APOE-Christchurch mutation into the three common APOE variants: APOE2Ch, APOE3Ch and APOE4Ch |
May 2, 2024 |
Mansour Haidar, Benjamin Schmid, Agustín Ruiz, et al. |
