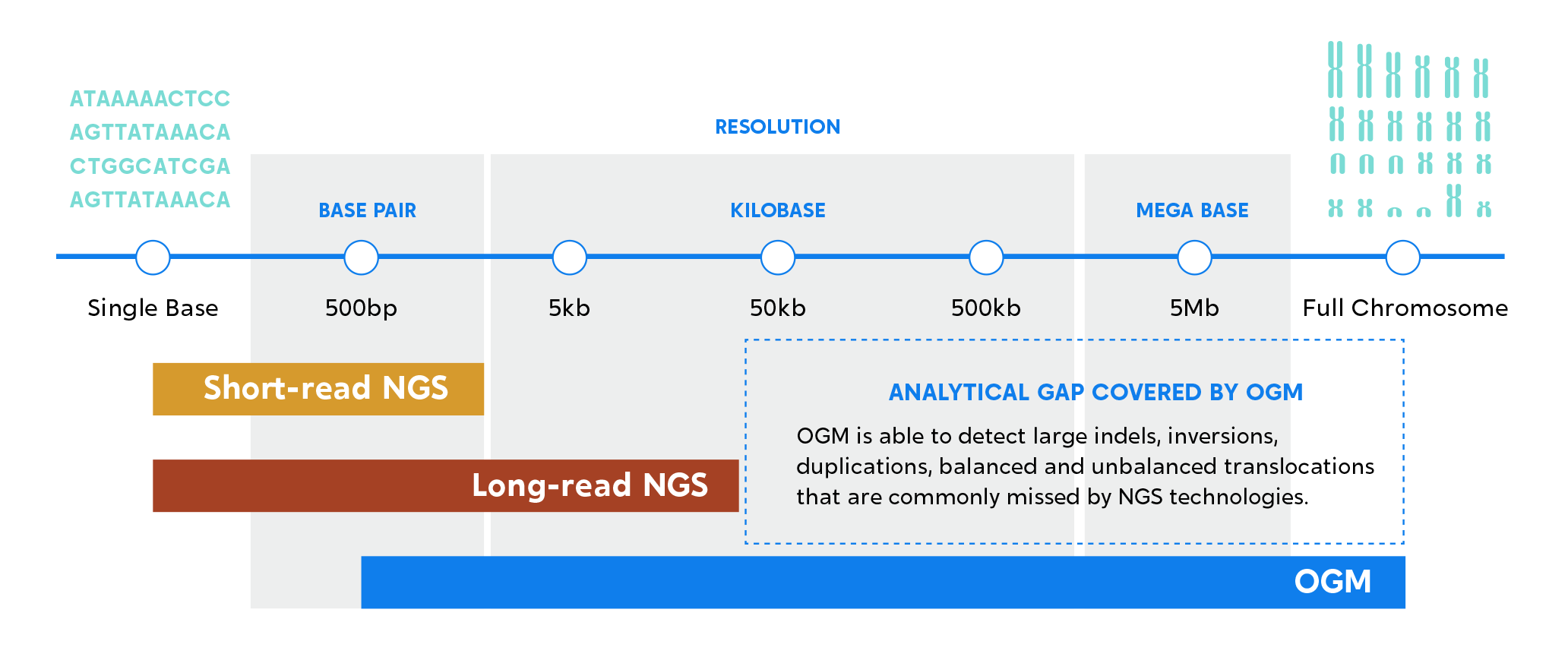
Structural variants (SVs) make up most of human genomic variation and are implicated in reproductive health issues and genetic diseases, such as infertility and miscarriages, neurodevelopmental delay, and rare diseases. The “gold standard” techniques used to identify pathogenic SVs linked to these diseases, including cytogenetic and molecular methods, often fall short.


Current approaches do not detect all potential SVs implicated in DD/ID etiology.11-12 Leverage the power of OGM to achieve results that correlate highly to traditional cytogenetic methods while revealing new pathogenic SVs in some cases where array and sequencing were unsuccessful.
OGM’s comprehensive SV detection empowers labs to increase their chances of finding pathogenic SVs that lead to rare genetic diseases.13-15


OGM is a tool that helps researchers better understand the underlying genetic etiology of recurrent miscarriage, infertility, and pregnancy abnormalities, including the ability to resolve some cryptic aberrations not detected with traditional methods.16-19
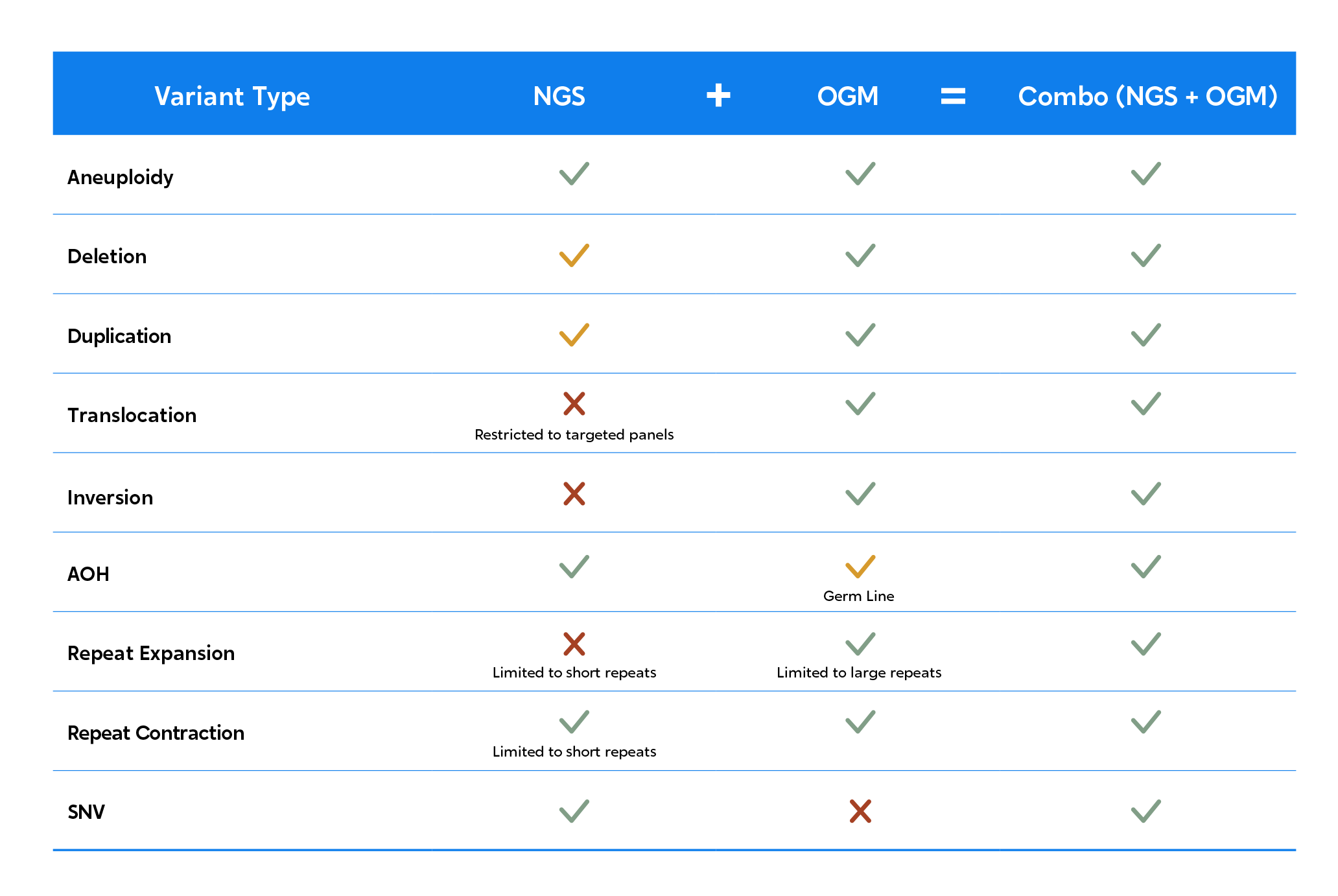
When OGM is added to the mix, you get better detection of insertions, deletions, duplications, translocations, inversions, and repeat expansions. Learn more about how OGM works
Previous studies have shown that a substantial portion of SVs detected by OGM are not detected by sequencing methods.20-22

OGM find new candidate genes
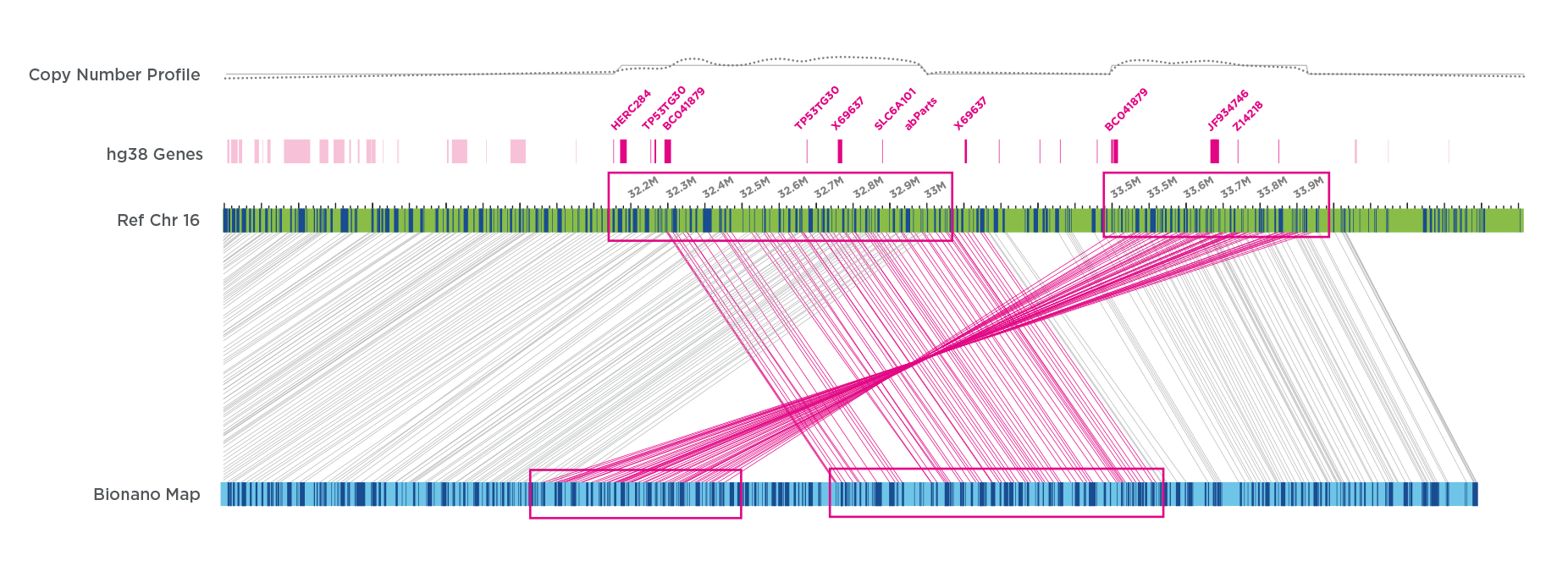
In a newborn with Congenital Diaphragmatic Hernia (CDH), a severe developmental disorder affecting the diaphragm, lungs and sometimes heart, OGM detected two adjacent duplications, one direct and one inverted. OGM revealed a much more complex architecture than could be inferred from microarray data and identified several additional candidate genes for CDH.24
OGM identifies new variants in known genes
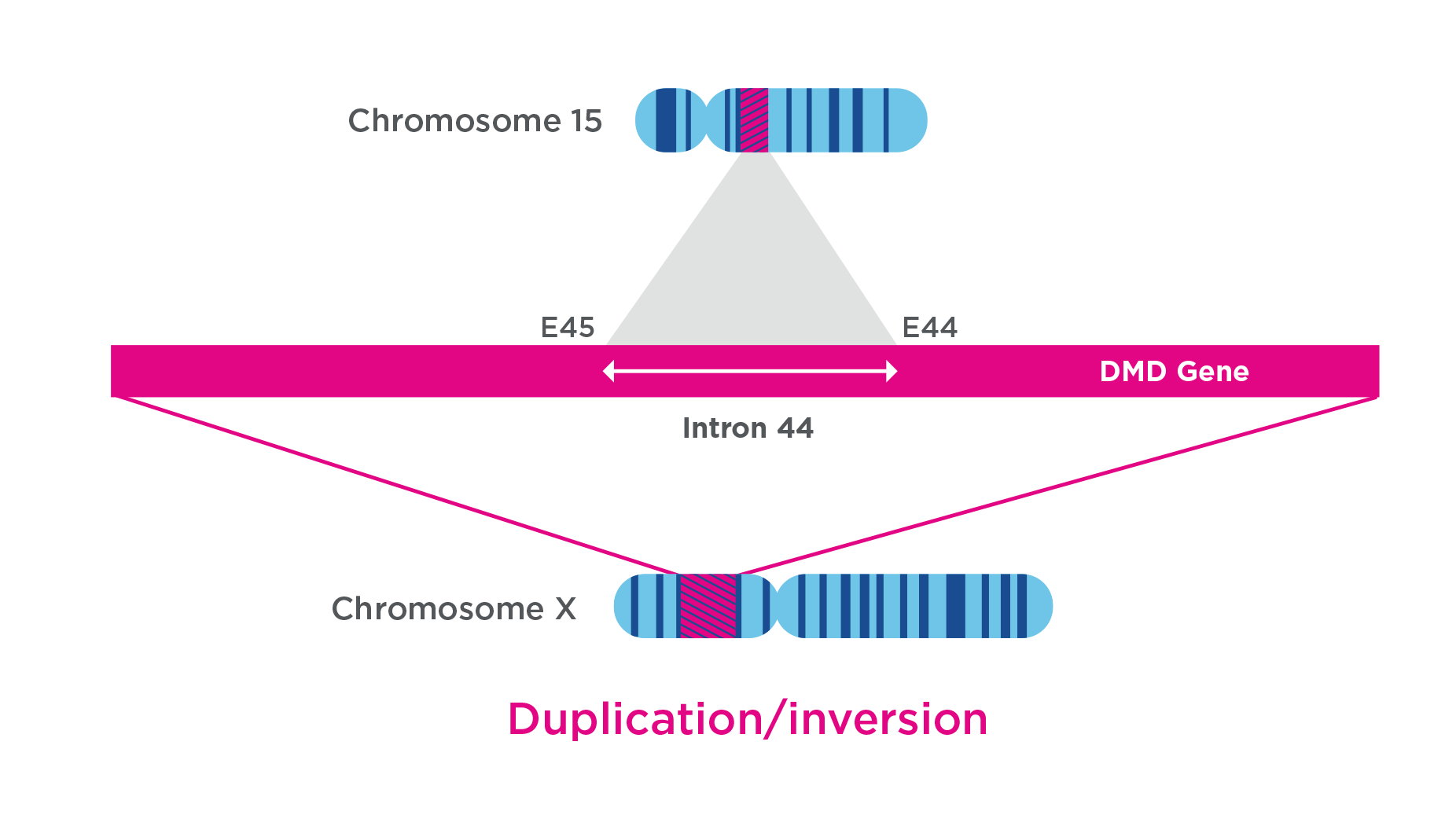
In a subject with Duchenne Muscular Dystrophy (DMD), a 420 kbp segment from chromosome 15 was duplicated in an inverted orientation in intron 44 of the Dystrophin gene. This insertion was not detected by NGS, and while chromosomal microarray can detect the duplication, its location and, therefore, implication in DMD could not be determined.25
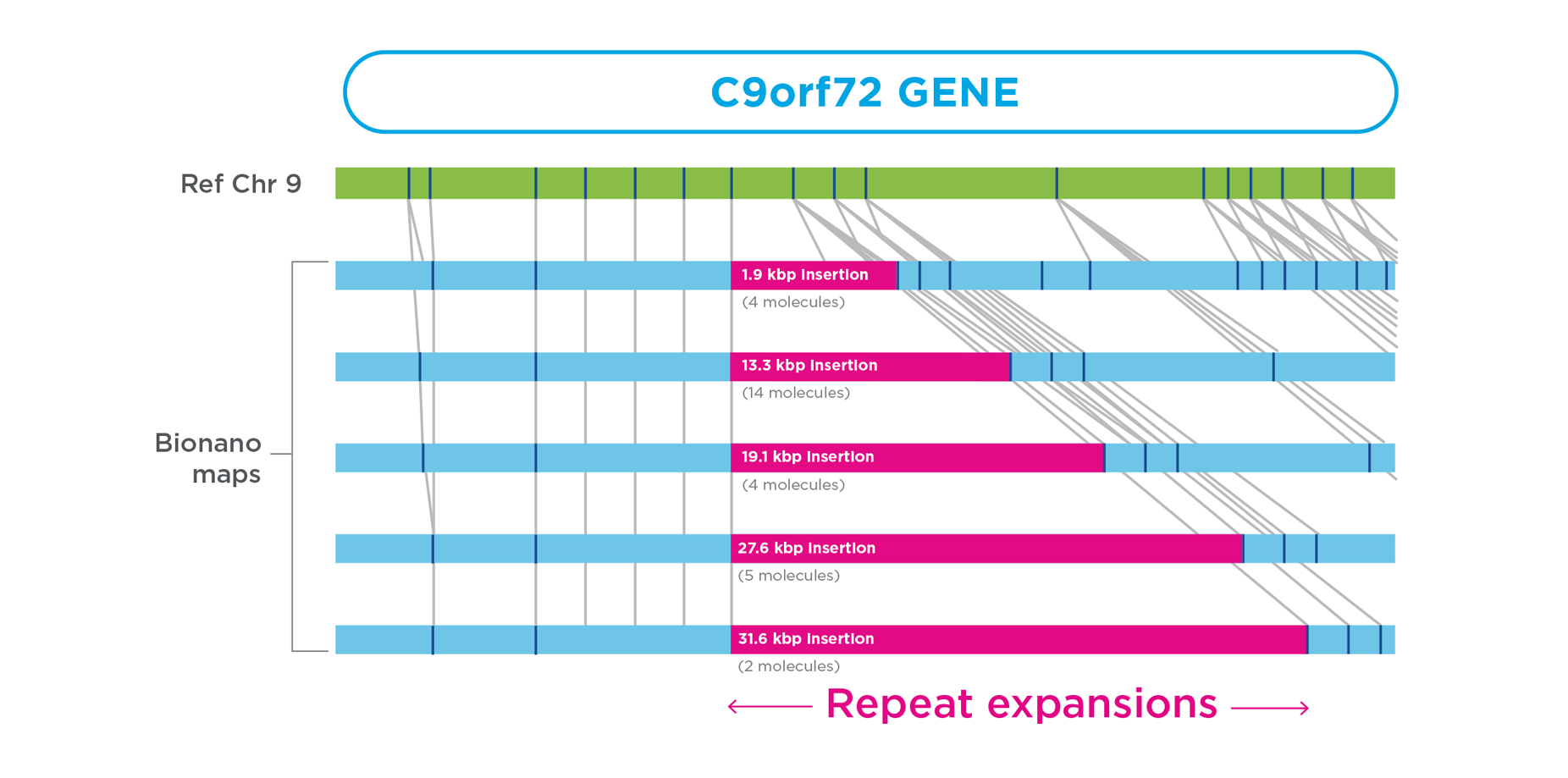
OGM reveals repeat expansions
In a single postmortem brain sample from an ALS subject, OGM detected a highly mosaic range of expansions of the C9orf72 GGGGCC repeat, ranging from the reference allele (not shown) to a 32 kbp expansion. No modern technology has been capable of spanning and measuring these large C9orf72 repeat expansions.26
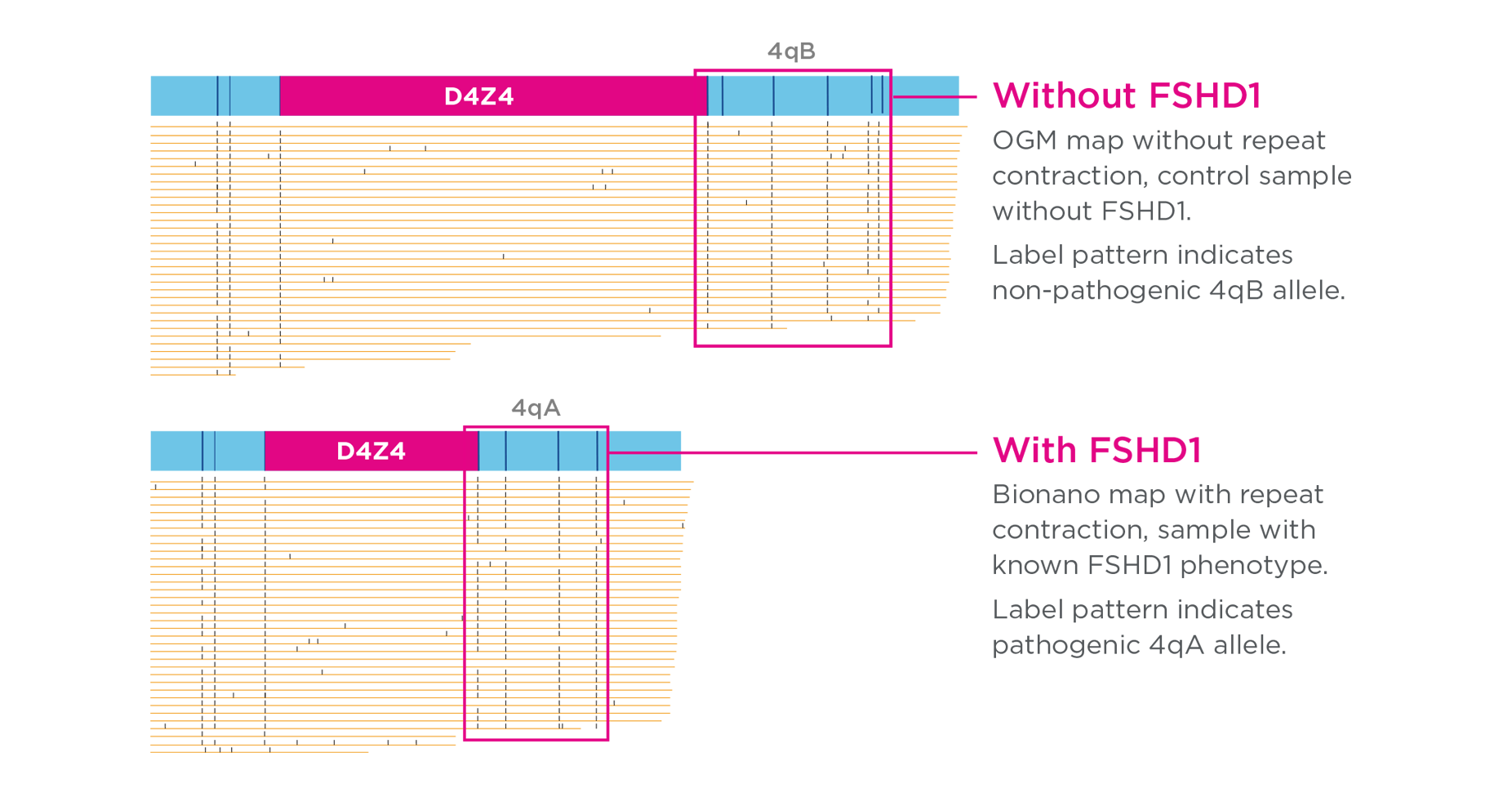
OGM detects FSHD1
Facioscapulohumeral Muscular Dystrophy (FSHD1) is a common form of muscular dystrophy with an extremely complex genotype. Correct genotyping requires the accurate sizing of a very large repeat region in the subtelomeric region of chromosome 4, a correct determining of the pathogenic vs non-pathogenic allele, and the distinction between the chromosome 4 repeat and an almost identical repeat on chromosome 10 not related to the disease. A team from the University of Iowa published the largest clinical research study to date evaluating OGM for FSHD1. The study, published in the Journal of Molecular Diagnostics, concluded that OGM can be performed more quickly, accurately, and reproducibly than the current gold standard method of Southern blot analysis.27
Check out expert presentations on the utility of OGM for evaluation of genetic disease
Watch VideosExplore the growing body of evidence for OGM performance in constitutional genetic disease in this downloadable publication summary.
DownloadRead about what structural variations are and why they matter.
Learn MoreSee how OGM reveals structural variation in a way that has never been done before.
Learn MoreFind the latest research in our Publications Library.
Learn More
| Title | Source | Authors |
|---|---|---|
| Optical genome mapping unveils hidden structural variants in neurodevelopmental disorders |
Nature May 16, 2024 |
Isabelle Schrauwen, Yasmin Rajendran, Anushree Acharya, et al. |
| A comprehensive approach to evaluate genetic abnormalities in multiple myeloma using optical genome mapping |
Blood Cancer Journal May 3, 2024 |
Ying S. Zou, Melanie Klausner, Jen Ghabrial, et al. |
| Generation of three isogenic, gene-edited iPSC lines carrying the APOE-Christchurch mutation into the three common APOE variants: APOE2Ch, APOE3Ch and APOE4Ch |
May 2, 2024 |
Mansour Haidar, Benjamin Schmid, Agustín Ruiz, et al. |
