Redefine Quality Control for Cell Bioprocessing with Optical Genome Mapping
Whether you are working with producer cell lines, research cell lines, or cell therapy applications, ensuring the genomic integrity and stability of your cell lines is of critical importance. Traditional cytogenetic methods have significant limitations in resolution, turnaround time, and scalability. Optical genome mapping (OGM) with the Bionano Saphyr® system is an advanced digital workflow that enables genome-wide analysis of structural variants (SVs) and copy number variants (CNVs), so you can easily screen cell lines for genomic instability and off-target events.


Screen your cell lines for all classes of structural variants in a single assay.
- OGM can detect aneuploidy, deletions, duplications, inversions, translocations, and repeat expansions or contractions
- Resolution 10,000x higher compared to traditional karyotyping
- Generate high-coverage data to detect low allele fractions (down to 5% with 400x coverage, currently available, and higher-coverage protocol under development)
Lower your costs and turnaround times by bringing the OGM assay in-house.
- Easy to implement in-house
- Consolidation of multiple tests drives cost savings
- Sample to answer in <1 week

Gain control over your cell QC operations.
- Fully digital workflow designed to scale
- Complete end-to-end solution including analysis software
- High-touch support from Bionano specialists
Advantages of Optical Genome Mapping Compared to Traditional Techniques
| Karyotype | Chromosomal Microarray | OGM | |
| Average TAT |
2+ weeks |
<1 week |
<1 week |
| Resolution |
5-10 Mbp |
>50-100 kbp |
>500 bp |
| Detects deletions and duplications |
(>5-10 Mbps) |
|
|
| Detects translocations and inversions |
(>5-10 Mbps) |
|
|
| Detects repeat instability | |||
| Scalable digital analysis | |||
| Easy to perform in-house |
Here’s How other labs have implemented OGM for Cell Line Screening and Quality Control
Redefine Quality Control for Cell Bioprocessing with Optical Genome Mapping
| Organization | Application | Methods | Findings |
|
CIRA Foundation (Japan) |
Evaluating the effects of CRISPR-Cas9 gene editing | Performed a stringent genomic integrity assessment of CRISPR-Cas9 edited IPSC subclones, using WGS, karyotyping and OGM | OGM uniquely identified unexpected chromosomal translocations and inversions introduced by gene editing |
| Oklahoma Medical Research Foundation (USA) | Evaluating the effects of prolonged cell culture on induced pluripotent stem cells (iPSCs) | Measured the effects of cell culturing in two iPSC lines in parallel for 50 passages and examined them at multiple time points using OGM | OGM identified substantial changes in the iPSC line genomes, including deletions, insertions, balanced translocations and inversions |
| Verve Therapeutics (USA)³ | Evaluating genomic integrity after (CRISPRcas) genome engineering in a primary liver cell line used in drug development | Assessed for chromosomal rearrangements and large insertions or deletions in a liver cell line treated with a single course gene editing drug in development | They showed that no additional SVs after treatment when accumulated compared with untreated controls |
| bit.bio (UK)* | Cytogenetic quality control of iPSCs | Assessed the cytogenetic health of IPSC banks at commercial scale | They adopted OGM in-house as a single workflow solution, replacing an outsourced two-assay process, reducing TAT from 5 weeks to 1 week and improving the quality of SV data |
References : 1. Kitano et al. Mol Ther Methods Clin Dev. 2022 : 26 : 15-25. doi : https://doi.org/10.1016/j.omtm.2022.05.010 2. Dubose et al. Genes. 2022; 13(7) : 1157. doi : https://www.mdpi.com/2073-4425/13/7/1157 3. Verve Therapeutics press release. April 26, 2022. https://ir.vervetx.com/news-releases/news-release-details/verve-therapeutics-presents-comprehensive-analyses-target 4. Bitbio website. May 5, 2022. https://www.bit.bio/blog/how-our-culture-enables-new-technologies
Discover How OGM Benefits Cell Bioprocessing Applications
Cytogenetic QC at bit.bio: How a next-generation cytogenetics platform enhances quality control of next-generation cells.
bit.bio
Watch Video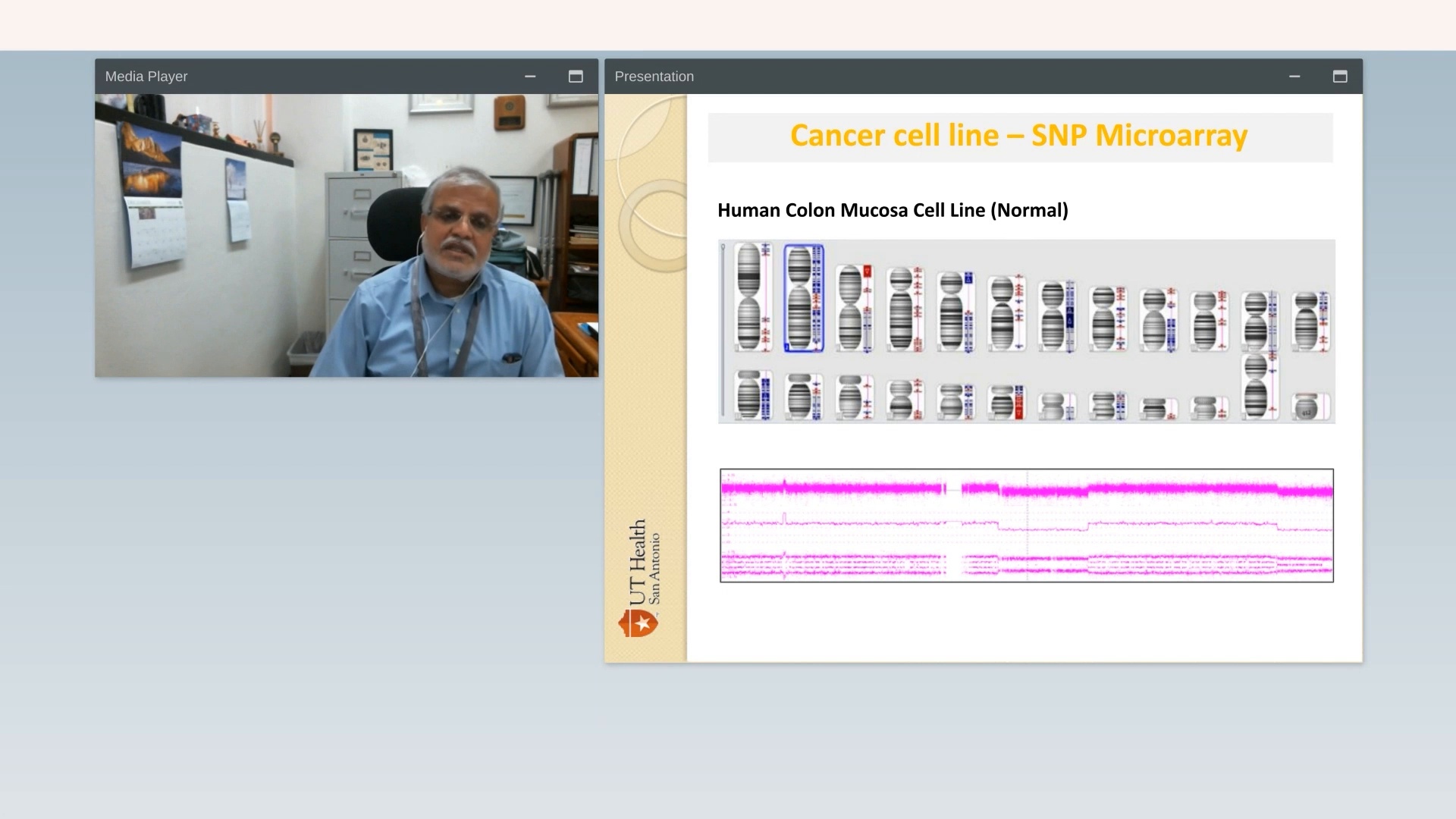
Vigor and reproducibility in research: The cell line saga: is OGM the answer?
UT Health – San Antonio
Watch VideoLearn More About OGM
Read about what structural variations are and why they matter.
Learn MoreSee how OGM reveals structural variation in a way that has never been done before.
Learn MoreFind the latest research in our Publications Library.
Learn More
Sign up to receive the latest research on OGM for cell bioprocessing applications.
Sign me up!Related Focus Areas & Applications
- Kitano et al. Mol Ther Methods Clin Dev. 2022; 26:15-25. doi: https://doi.org/10.1016/j.omtm.2022.05.010
- Dubose et al. Genes. 2022; 13(7):1157. doi: https://www.mdpi.com/2073-4425/13/7/1157
- Verve Therapeutics press release. April 26, 2022. https://ir.vervetx.com/news-releases/news-release-details/verve-therapeutics-presents-comprehensive-analyses-target
- Bit.bio website. May 5, 2022. https://www.bit.bio/blog/how-our-culture-enables-new-technologies



