Detecting broken, missing, rearranged, or extra chromosomes provides critical, actionable insights for cancer and genetic diseases, yet traditional cytogenetic methods fall short of discovering key chromosomal aberrations.





Reveal all chromosomal aberrations commonly detected by classical cytogenetics while significantly increasing pathogenic findings with a simpler workflow.
Multiple studies at renowned institutions worldwide show the same outcomes. OGM not only has a high correlation to results from traditional methods but also reveals additional actionable pathogenic abnormalities.1-8
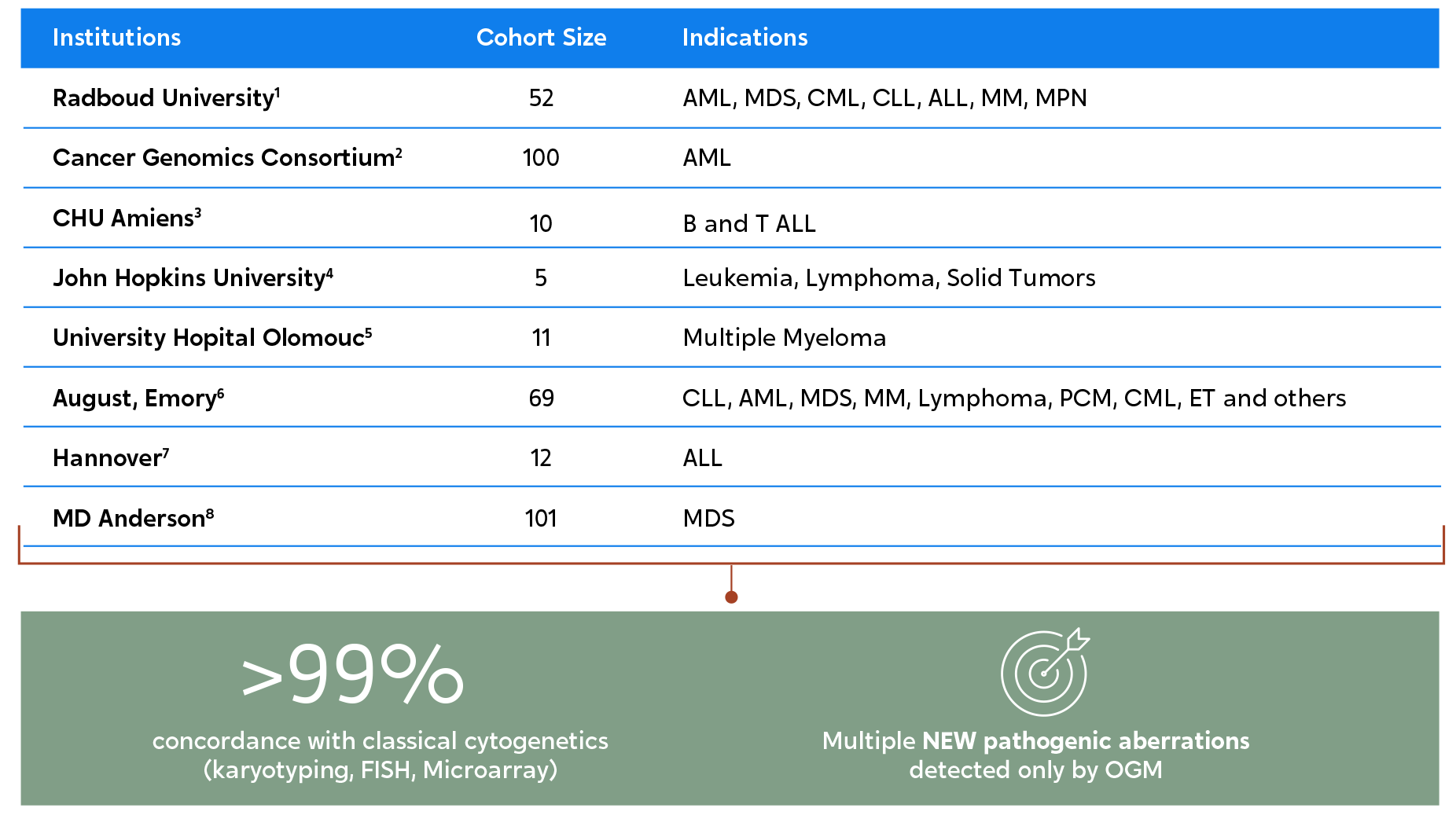
European multi-site consortium, aimed at evaluating OGM’s performance as compared to classical methods (microarray and karyotyping), across 85 constitutional samples. Nearly 100 chromosomal aberrations were evaluated, many mapping to complex regions of the human genome, and involved in repeat-mediated microdeletion/microduplication syndromes.
Institutions involved:
Author’s Conclusion:
“These results highlight the potential for OGM to provide a cost-effective and easy-to-use alternative that would allow comprehensive detection of chromosomal aberrations and structural variants, which could give rise to an era of “next-generation cytogenetics.”
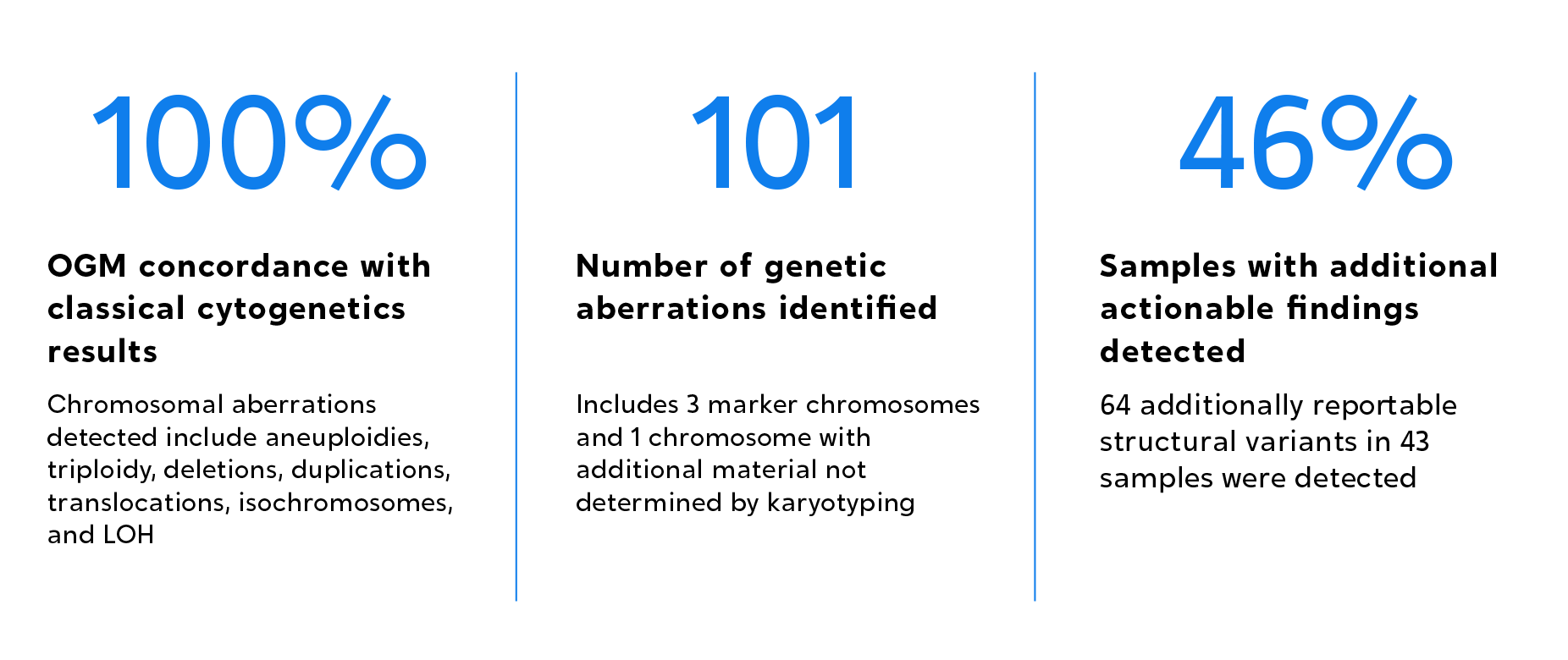
OGM was proven to detect multiple categories of SVs with 100% concordance compared to classical methods.
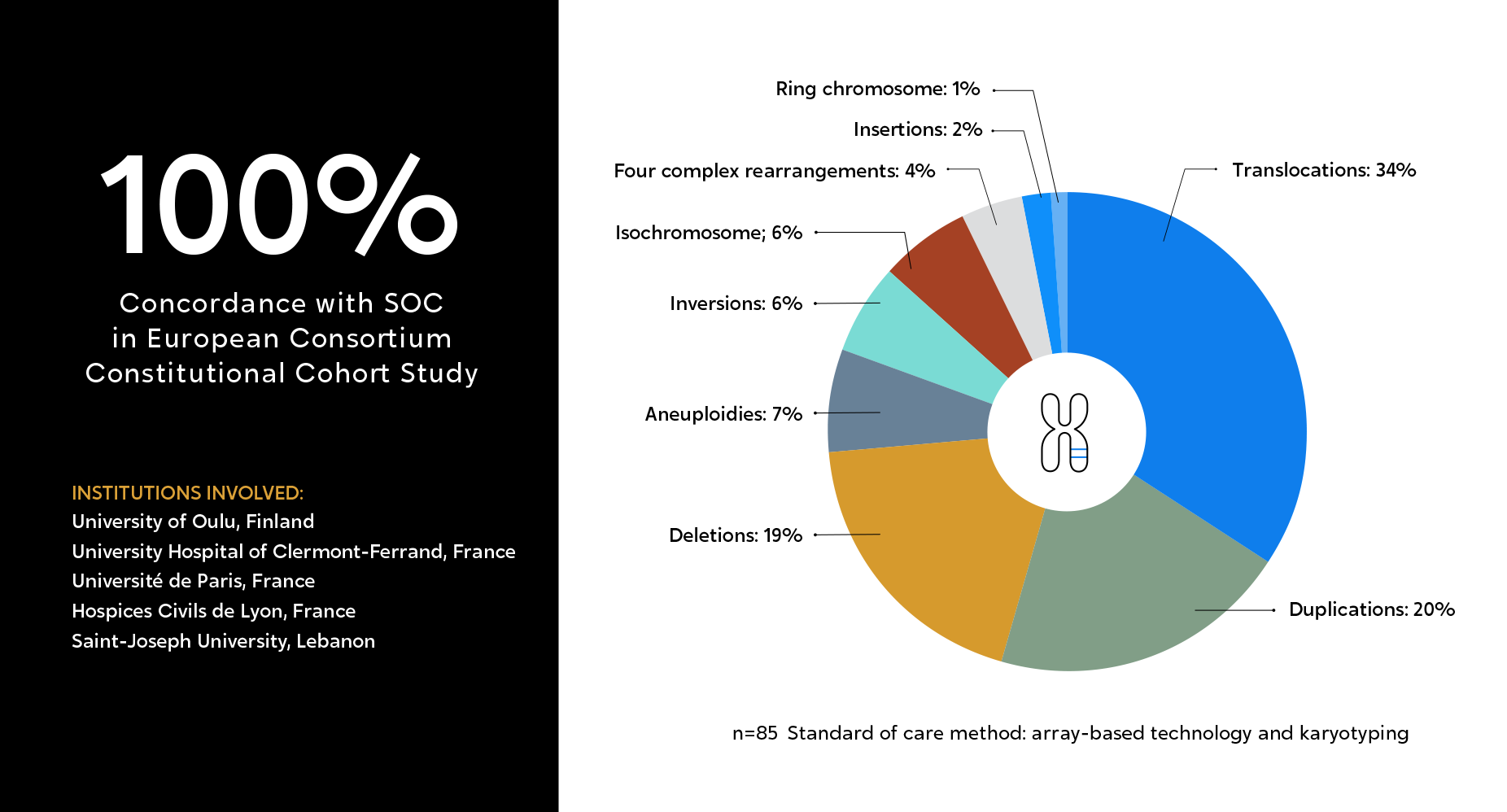
American multi-center, double-blinded study, aimed to evaluate OGM’s performance, as compared to classical methods (karyotyping, microarray, and FISH), across 331 constitutional samples.
Institutions involved:
Outstanding performance for OGM across all types of SVs evaluated.
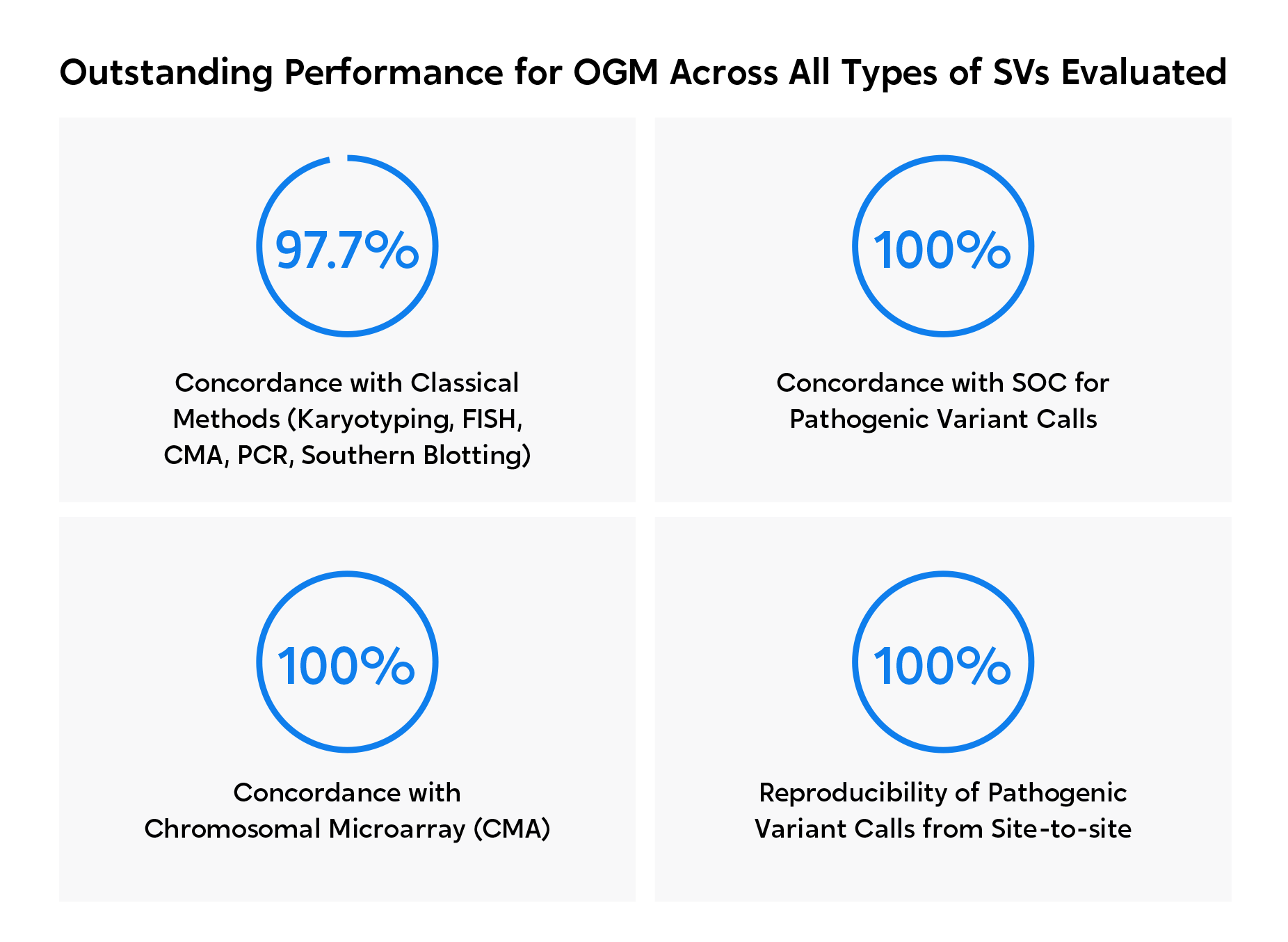
OGM was proven to not only achieve 100% concordance with classical cytogenetic methods, but also to unveil new pathogenic aberrations in 46% of a total of 94 amniocentesis samples evaluated.
Author’s statement:
“We report the feasibility and relative ease of implementing OGM for prenatal assessment compared to classical methods with the platform, demonstrating high robust technical and analytical performance and recommend OGM as a first-tier assay in prenatal settings.”
Large-scale study shows 100% concordance between OGM and cytogenetics in prenatal cohort with additional findings.
Learn how OGM meets the challenges of new blood cancer classifications.
Featuring Dr. Adam Smith, University Health Network, Toronto, Ontario, Canada
Watch On-demandHear perspectives from a pathologist and an oncologist on the utility of OGM for evaluation of myelodysplastic syndromes.
Featuring Dr. Rashmi Kanagal-Shamanna and Dr. Guillermo Garcia-Manero, MD Anderson Cancer Center
Watch On-demandRead about what structural variations are and why they matter.
Learn MoreSee how OGM reveals structural variation in a way that has never been done before.
Learn MoreFind the latest research in our Publications Library.
Learn More
| Title | Source | Authors |
|---|---|---|
| Optical genome mapping unveils hidden structural variants in neurodevelopmental disorders |
Nature May 16, 2024 |
Isabelle Schrauwen, Yasmin Rajendran, Anushree Acharya, et al. |
| A comprehensive approach to evaluate genetic abnormalities in multiple myeloma using optical genome mapping |
Blood Cancer Journal May 3, 2024 |
Ying S. Zou, Melanie Klausner, Jen Ghabrial, et al. |
| Generation of three isogenic, gene-edited iPSC lines carrying the APOE-Christchurch mutation into the three common APOE variants: APOE2Ch, APOE3Ch and APOE4Ch |
May 2, 2024 |
Mansour Haidar, Benjamin Schmid, Agustín Ruiz, et al. |
