Structural variants (SVs) are a hallmark of cancer, yet next-generation sequencing (NGS) methods fail to detect all classes and sizes of SVs, missing a significant amount of information critical to understanding cancer biology.1-4 Studies have shown that both short-read and long-read sequencing technologies miss a significant portion of SVs detected by optical genome mapping (OGM).1,2
Combine NGS with OGM to unlock a broader spectrum of genetic variants, generate a more complete cancer genome profile, and discover new actionable biomarkers.




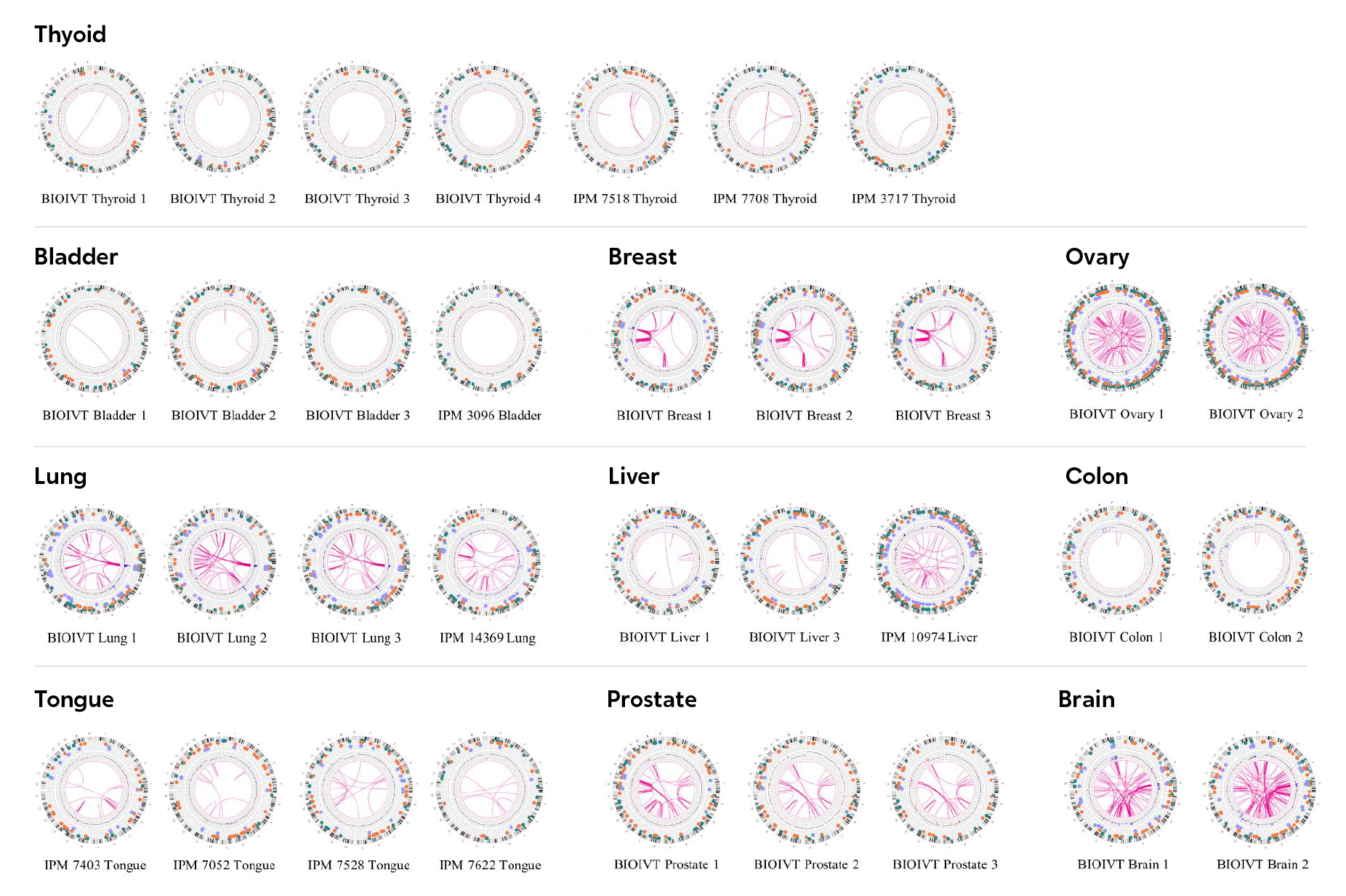
Study with Multiple Tumor Types Reveals SVs That Would Not Be Readily Identified by Targeted Gene Panels Generally Used to Assess Tumor Genomes:
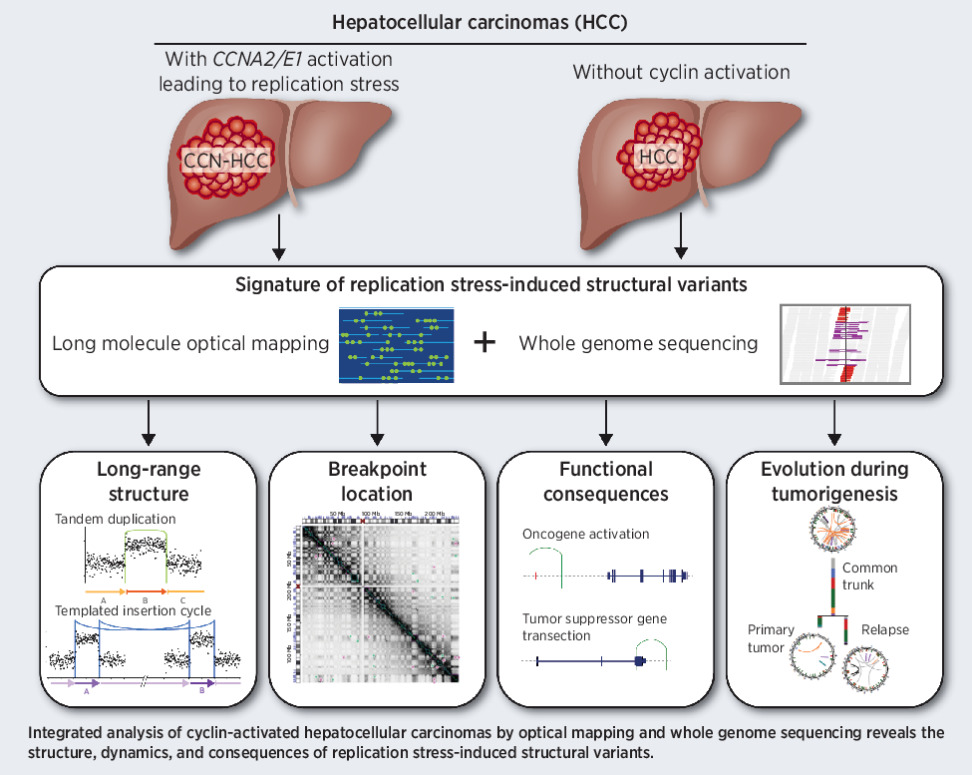
By combining WGS and optical mapping, researchers could reconstruct the structure of complex SVs at large scale and single-base resolution in a subgroup of hepatocellular carcinoma with cyclin-induced replication stress.1
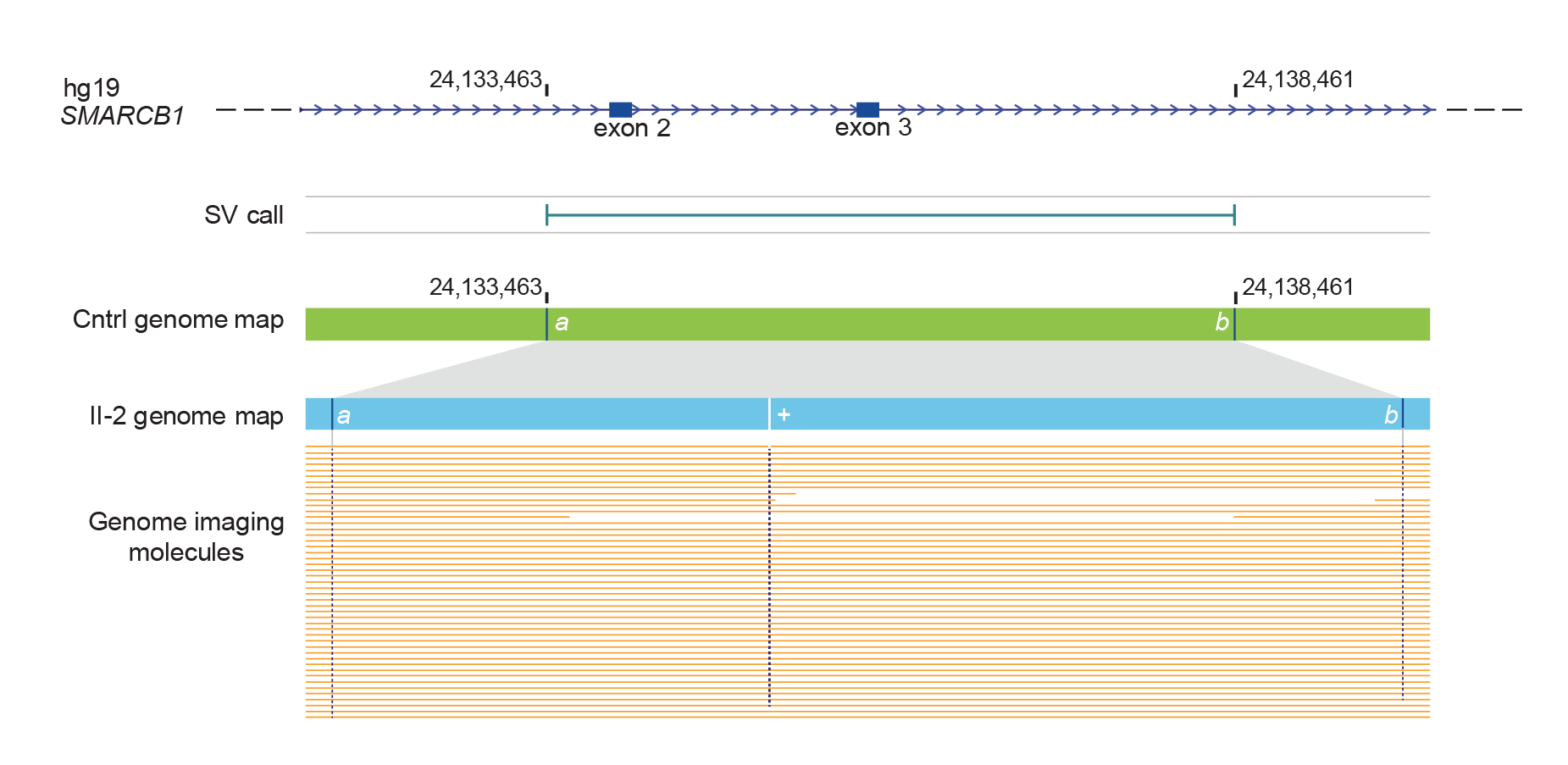
OGM Solved Cancer Predisposition Mystery in a Pediatric ATRT Case
Previous analysis using Sanger sequencing, NGS (whole exome and whole genome sequencing), and MLPA could not identify a causative variant.
OGM revealed a 2.7kb insertion in the ATRT gene.4 OGM analysis of germline DNA showing an SV call enclosing the insertion at the SMARCB1 locus.
Utility of optical genome mapping for the chromosomal characterization of solid tumors.
Ravindra Kolhe, MD, PhD, FCAP
Optical genome mapping reveals novel structural variants in pediatric brain tumors.
Miriam Bornhorst, MD
Read about what structural variations are and why they matter.
Learn MoreSee how OGM reveals structural variation in a way that has never been done before.
Learn MoreFind the latest research in our Publications Library.
Learn More
| Title | Source | Authors |
|---|---|---|
| Optical genome mapping unveils hidden structural variants in neurodevelopmental disorders |
Nature May 16, 2024 |
Isabelle Schrauwen, Yasmin Rajendran, Anushree Acharya, et al. |
| A comprehensive approach to evaluate genetic abnormalities in multiple myeloma using optical genome mapping |
Blood Cancer Journal May 3, 2024 |
Ying S. Zou, Melanie Klausner, Jen Ghabrial, et al. |
| Generation of three isogenic, gene-edited iPSC lines carrying the APOE-Christchurch mutation into the three common APOE variants: APOE2Ch, APOE3Ch and APOE4Ch |
May 2, 2024 |
Mansour Haidar, Benjamin Schmid, Agustín Ruiz, et al. |
