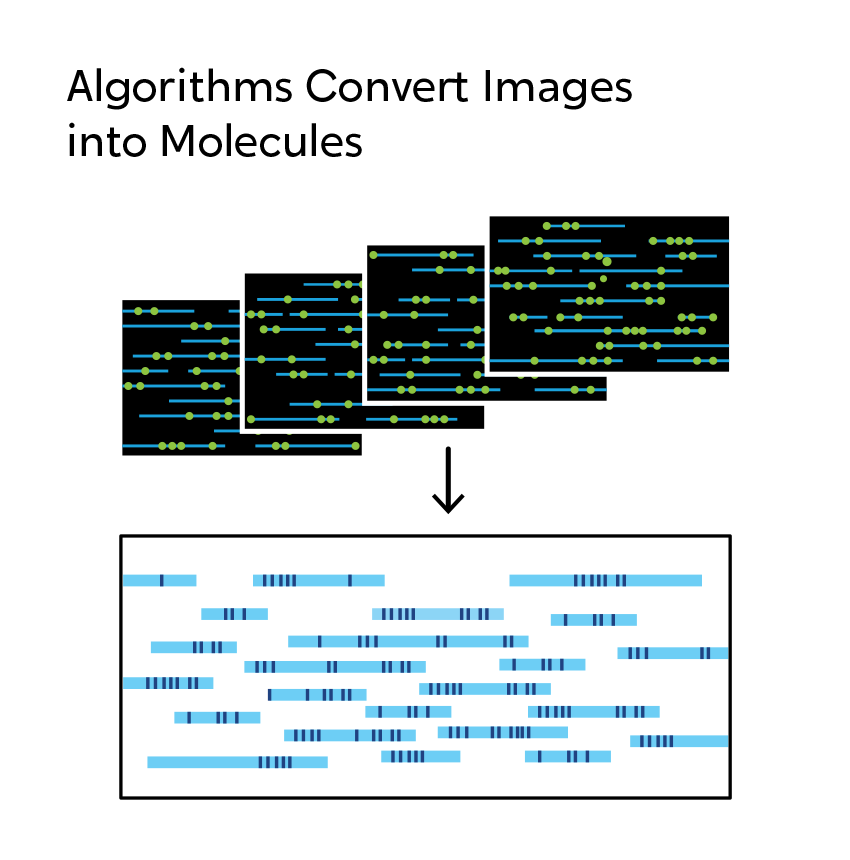
An Overview of OGM
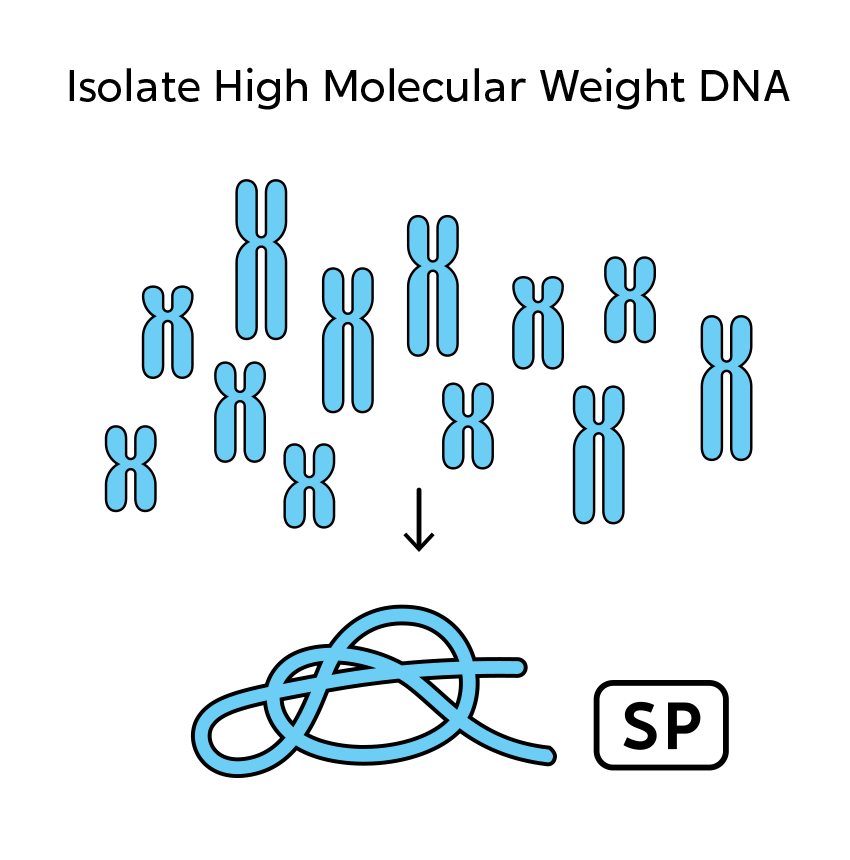
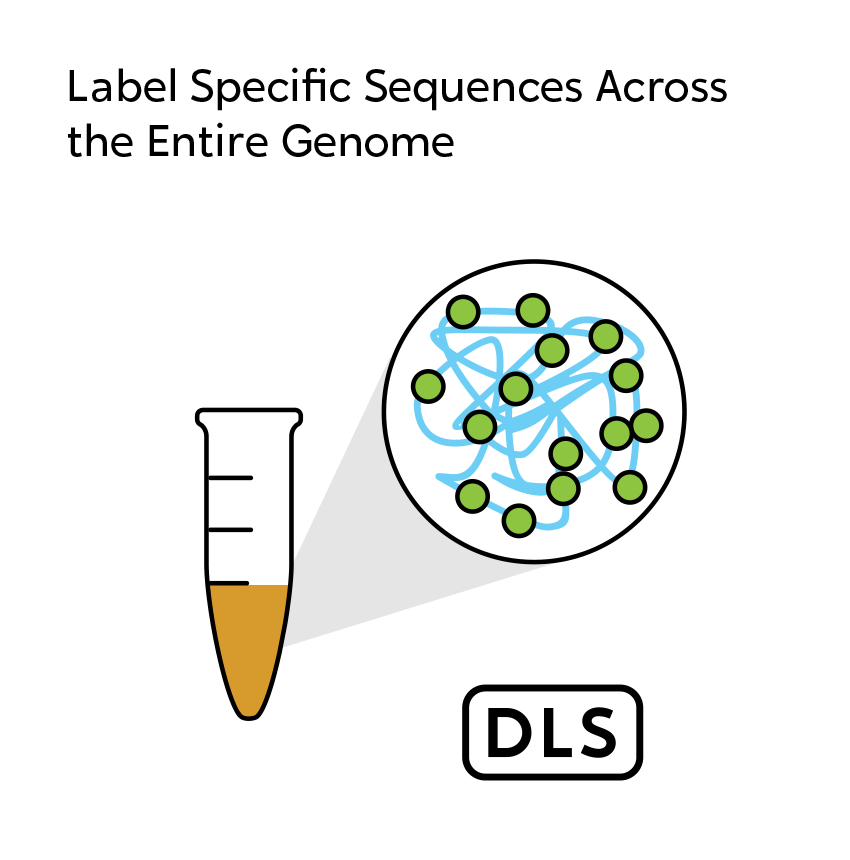
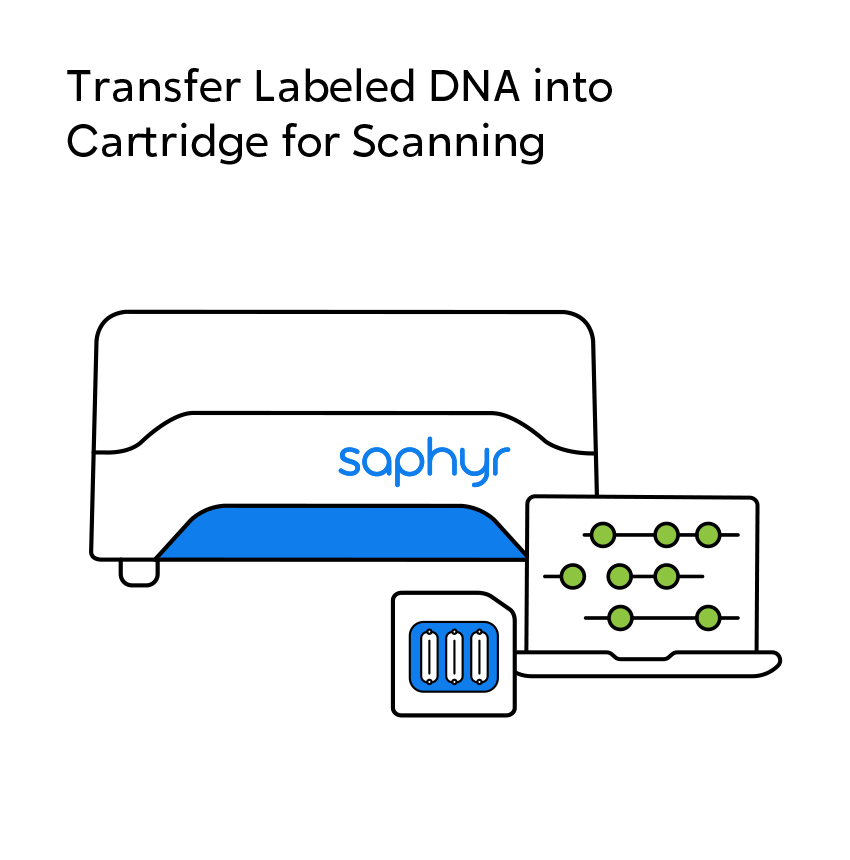
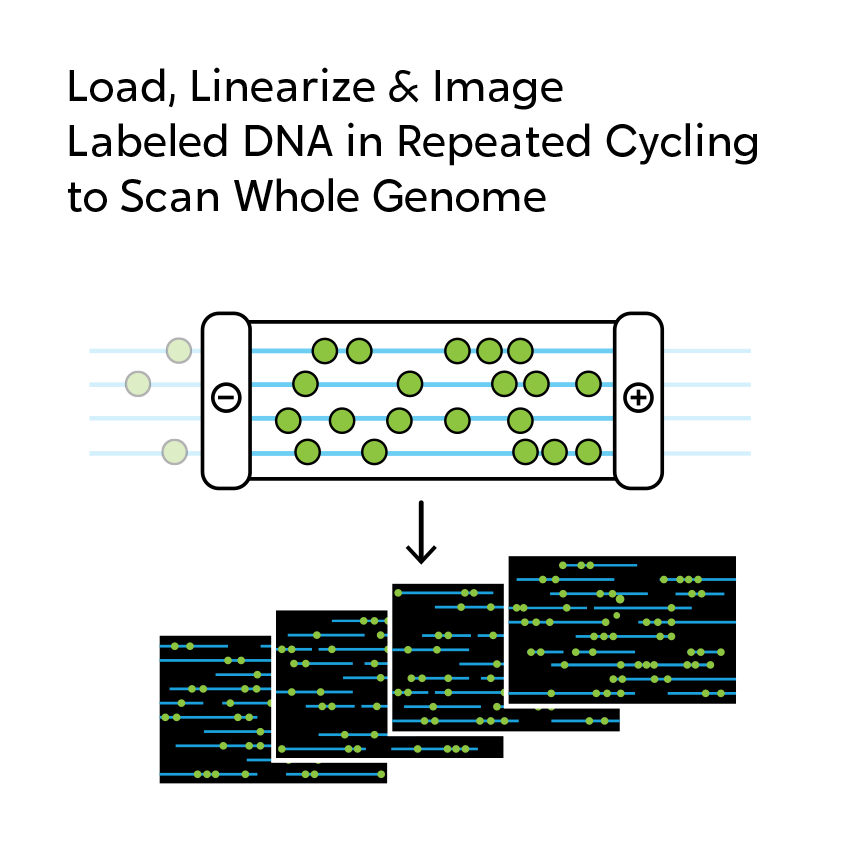
OGM begins with the isolation of ultra-high molecular weight (UHMW) DNA from blood, bone marrow aspirates, cultured cells (including chorionic villi and amniocytes), tissue, or tumor biopsies. A single enzymatic reaction places fluorescent labels all throughout the genome at a specific sequence motif. The labeled DNA molecules are linearized in nanochannel arrays on the chip. Changes in the patterning or spacing of the labels are identified by software solutions to accurately detect all classes of structural variants (SVs). The OGM data generated can be analyzed alone, or in combination with sequencing or array data.

Observe SVs Directly with OGM
When using OGM, structural variations are observed, not inferred as they are with next-generation sequencing (NGS). When short-read NGS sequences are aligned to reference genomes, megabase-size native DNA molecules are imaged, and most large structural variations or their breakpoints can be observed directly in the label pattern on the molecules. In addition, SVs can occur in the repetitive sequences that make up two-thirds of the genome. NGS has significant, known limitations with assessing variation in and around repetitive sequences. OGM is free from these constraints, making it possible to reveal those changes that can contribute to human disease.

Related Applications
Learn More about OGM
Read about what structural variations are and why they matter.
Learn MoreFind the latest research in our Publications Library.
Learn MoreLearn from a specialist: Request a live overview of how OGM works and discuss how your research can benefit. Request an OGM overview
Request an OGM Overview