Blog
OMIM data in NxClinical
OMIM (Online Mendelian Inheritance in Man) is a comprehensive database consisting of human genes and genetic phenotypes associated with Mendelian disorders. The database is used by genetic labs to assist in interpretation. It is maintained by Johns Hopkins University and is available freely via the online GUI for personal, educational, or research use but other uses, usage by certain types of organizations, as well as downloading or incorporating with products does carry licensing fees.
BioDiscovery licenses OMIM and provides the content within NxClinical software so you have access to the OMIM data directly within the software and you don’t have to pay for a separate license. We download and curate the OMIM content and transform the data into useful tracks to assist you in your interpretation process.
OMIM tracks can be used in different ways within NxClinical. The tracks can be added to the genome browser in NxClinical and events here are hyperlinked to the OMIM page for each event. It can also be used to automatically pre-classify events following lab specific and/or regional guidelines (e.g. ACMG).
To enhance the utility of OMIM content, we curate and divide it into several tracks including the following:
- OMIM Genes – all the genes represented in OMIM
- OMIM Morbid Map – all genes associated with a disorder in OMIM
- OMIM Morbid Phenotypes – phenotypes associated with a disorder in OMIM; the track shows the cytogenetic location, gene symbol, phenotype, inheritance (if available) and the phenotype mapping key.
- Phenotype mapping key values from OMIM:
- (1) the disorder was positioned by mapping of the wildtype gene
- (2) the disease phenotype itself was mapped
- (3) the molecular basis of the disorder is known
- (4) the disorder is a chromosome deletion or duplication syndrome
- Phenotype mapping key values from OMIM:
All features displayed in the tracks in the genome browser are hyperlinked to the respective OMIM page.
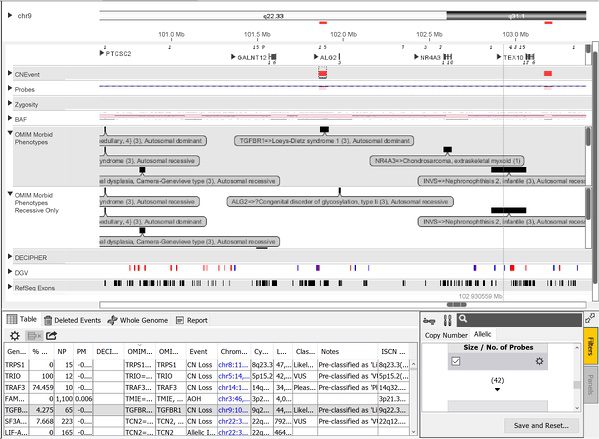
The OMIM content described above is also further partitioned into additional categories based on phenotype. Use of these is especially useful for filtering and in the automated pre-classification system. This system uses a set of user-defined rules (can be those recommended by ACMG) to “pre-classify” events thereby greatly reducing the amount of time a reviewer needs to spend with each case. Once the events are pre-classified automatically, the reviewer can manually look at the more difficult-to-interpret events. Events can be pre-classified based on a number of criteria including how the event was classified within the lab’s historical cases, whether the event overlaps exons/specific genes, how it is categorized in other databases such as DECIPHER and OMIM, etc. For the OMIM Morbid Phenotypes data, the content is further divided into additional tracks as depicted below.
Dominant:
- Autosomal Dominant
- X-linked Dominant
Recessive:
- Autosomal Recessive
- X-linked Recessive
Duplication Syndromes
Deletion Syndromes
For example, when performing carrier screening on the mother, using the Decision Tree one can automatically flag a loss event that overlaps one in the X-linked Recessive list as potentially pathogenic since a male child would have 50% chance of having the condition. The ability to identify and mark such events appropriately in an automated fashion greatly facilitates the review process and having such segregated tracks is very in this process.
OMIM is a great resource to use during the interpretation process and is provided for free with NxClinical offering substantial savings for a lab. In addition, the data is curated by BioDiscovery and sorted into different lists to further enhance the utility of this great data set.



