Blog
A first-of-its-kind clinical research genetics knowledgebase – Accelerate your case review with NxClinical
Conventional genomic analysis software has, for years, forced frustrating inefficiencies on clinical labs tasked with reviewing constitutional and somatic patient samples and making critical time-sensitive decisions.
Two software shortcomings, in particular, tend to hurt the speed and quality of the clinical investigation and reporting that comprises these reviews, especially with complex cases:
- First, conventional software doesn’t help labs build and use their own reported interpretations from historical cases. Similarly, most software doesn’t integrate directly with third-party databases for comparing similar genomic aberrations and profiles. Without an integrated tool to query published clinical evidence for similar variants, users are forced to scour homegrown note-taking systems and disparate online databases to handle annotation on their own.
- Second, current software designs limit any clinically relevant information that users do find to a systematic manual annotation process for each case; increasing turnaround time and inviting inconsistency. Once users collect some existing “knowledge” about a matching genomic aberration—for example, a particular syndrome and associated penetrance—typical software solutions force them to awkwardly work across multiple systems to report on those findings, completely “by hand”. Without a convenient interface, the format and quality of that reporting can therefore vary widely from case to case.
Taken together, the hands-on-time required to gather clinical evidence and report on patient cases can represent one of the most frustratingly time-consuming processes in a genetic lab’s workflow.
In an October 2021 article published in The Journal of Molecular Medicine, members of an Association for Molecular Pathology working group focused on electronic health record (EHR) interoperability for clinical genomics data detailed many of the issues with institutions not having a place to store variant data, stating, “[t]he single most important architectural component that would facilitate the use of genomic data in EHRs is precisely what most, if not all, EHRs lack: the ability to consume and store individual variants discretely in the database in a scalable, standardized, and reliable way.”
The newest version of NxClinical fills this critical role—and solves the two conventional software problems identified above—with its integrated Knowledgebase (KB) module. This first-of-its-kind database feature can dramatically simplify a lab’s interpretation, reporting process, and standardization while enhancing the consistency of each case report.
At a glance, the KB stores information about a variant, a genomic region, or a genome-wide signature, allowing users to create a database of syndromes, cytogenetic disorders, pathogenic variants, or cancer genomic profiles, tailored to their specific methodology. In fact, NxClinical allows for multiple KBs, designed either for postnatal or oncology workflows, and is split between CNV, Seq Var, and AOH events, whether focal or as genome-wide patterns.
- For constitutional cases, users can browse through annotations composed of causative gene lists, inheritance mode, event interpretation, PubMed references, evidence level, and phenotypes.
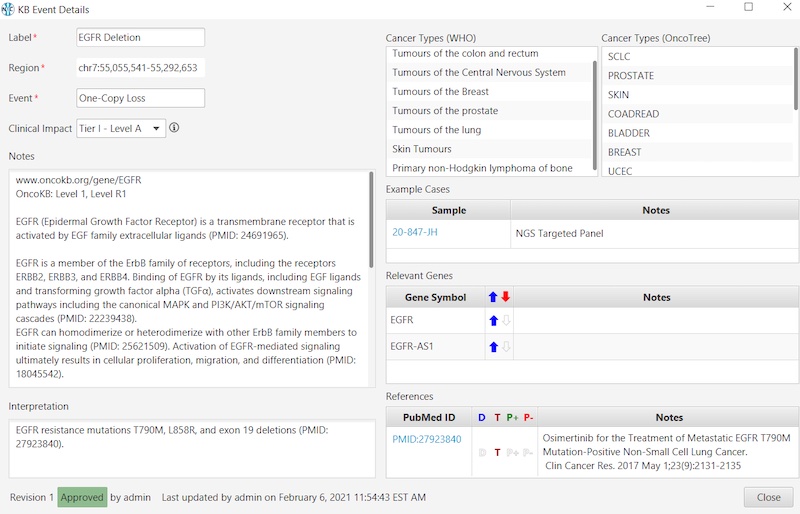
- For oncology cases, the KB stores cancer-specific information such as cancer type from the WHO and OncoTree ontologies, gene functions (oncogene/tumor suppressor), and AMP-ASCO-CAP recommended classifications. The KB also comprises relevant PubMed references, summarized for their impact on diagnostic, therapeutic, and prognostic outcomes.
Interested in seeing first-hand how much the NxClinical’s Knowledgebase can streamline and strengthen your lab’s constitutional and oncology workflows? Request a free personalized demo. We’ll connect on an initial consultation to answer any questions and dive a little deeper before demonstrating NxClinical with example data or your own.
Want to learn more before reaching out to explore NxClinical further? Head to our product update page for a wealth of information on the latest version of our software or read on to learn more about how NxClinical integrated Knowledgebase solves a number of long-standing problems in today’s clinical workflows.
Free tutorial for NxClinical users
Making your oncology case review process more efficient through an institutional knowledgebase integrated with NxClinical 6.0
Are you an active NxClinical user considering an update? In this 25-minute webinar, Soheil Shams, Founder & CEO of BioDiscovery, uses multiple example oncology cases to demonstrate the most effective workflow and case review benefits of the Knowledgebase in NxClinical 6.0.
The costly inefficiencies of operating in a non-integrated knowledge ecosystem
Most cytogenetic labs operate in non-integrated knowledge ecosystems—meaning they don’t have a single software platform for building a database of clinical evidence and must typically gather and curate collated information from querying each patient findings against multiple online sources (such as ClinGen, dbSNP, DECIPHER, DGV, GHR, gnomAD, HGMD or OMIM).
For those equipped with some knowledge ecosystems, the solution is usually fragmented at best (i.e. information saved as snippets)—and non-existent at worst. If any user can make insightful data comparisons at all, it often comes at a high cost of time, finding information from online resources, filtering the clinical evidence for matching methodologies (see example below), and manually crafting a relevant report. As we’ve heard from labs firsthand, this drag on efficiency can be significant.
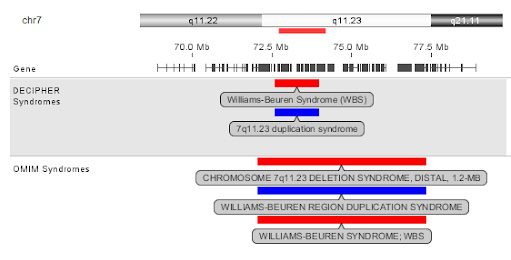
Until recently, an integrated knowledgebase function that enabled and automated this work simply didn’t exist in any software typically found in clinical labs to analyze and interpret genomic variants from microarray and NGS data. Without this capability, labs have had to devote a significant amount of time looking back through previous case notes, spreadsheets, or other ad hoc databases to manually locate similar case data, reconcile annotations, and run comparisons to inform their decisions and reporting.
These manual solutions are both inefficient and unscalable. Non-integrated knowledge systems and processes can be disrupted or completely lost upon staff turnover. Inefficiencies and inconsistencies are often amplified from multiple users’ input (for instance with irregularities from note-taking). In clinical settings obligated to comply with standards and operating protocols, the net result is always the same: slower turnaround time, inconsistent reporting, and lack of standardization.
A dedicated clinical lab knowledgebase centralizes that ecosystem—accelerating all of these processes and improving the reliability of the analysis and reporting…
How NxClinical’s Knowledgebase makes case review workflows more efficient and scalable
Clinical genetics labs have long-deserved an analysis software that prevents them from such aforementioned challenges. Labs should be able to collect, organize, validate annotations and interpretations in a seamless and standardized manner. Moreover, genome analysts and directors should be able to access this curated information for any CNV, Seq Var, AOH event, or genome-wide profile, and rapidly validate their decision making accordingly.
NxClinical’s KB does just that as an integrative component of each patient review workflow.
The KB enables labs to compile a comprehensive set of clinically-useful reference information and quickly find and access that information directly from within NxClinical. Without having to switch between systems, users can rapidly address the causative effect of a patient’s phenotype, or validate a cancer diagnosis, therapeutic option, and prognosis outcome from a genome-wide profile. Labs can use the KB to consolidate curated evidence from various sources gathered over time, thereby developing a more compliant, robust, and sustainable case review process.
The KB was explicitly developed to fill the gap in software capabilities and addresses recurrent feedback from many cytogenetic consortia. It’s been designed and integrated into NxClinical specifically to support complex constitutional and oncology case review, often driven by genome-wide CNV or AOH signatures—all to enable the best-informed decisions while assisting labs with compliance and performance in due time.
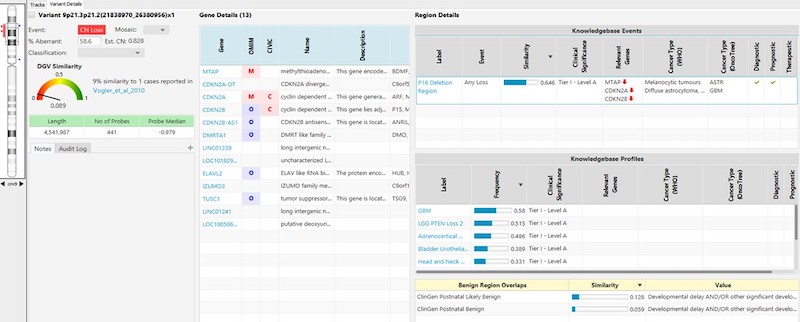
- A simple, standardized entry form enables users to quickly and easily submit information into the KB. (See example entry forms below.)
- A robust approval system can be configured to tightly regulate entries and ensure that only complete and accurate information enters the KB. (This also helps alleviate the long-standing problem of relying solely on curated public databases that can contain untrustworthy data.)
- The KB supports two distinct types of applications—“Oncology” and “Constitutional”—each containing fields unique to the specified application type.
- For oncology, the KB within NxClincial provides users with a global similarity score—a statistical analysis on the degree to which a patient sample matches a cancer pattern or a cancer profile in the database.
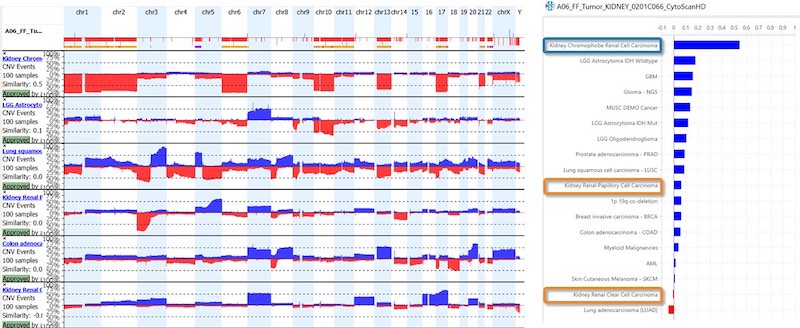
- In addition to confirming a diagnosis, the KB can also help to infer the prognosis by providing clinically relevant data suggesting how well a patient may respond to a given therapy.
- The most recent release of NxClinical offers a new overlay of sequence variants and uniparental disomy. These two types of aberrations can also be entered into the KB.
To illustrate the simplicity and convenience for the user, here’s an example of a Constitutional KB entry:
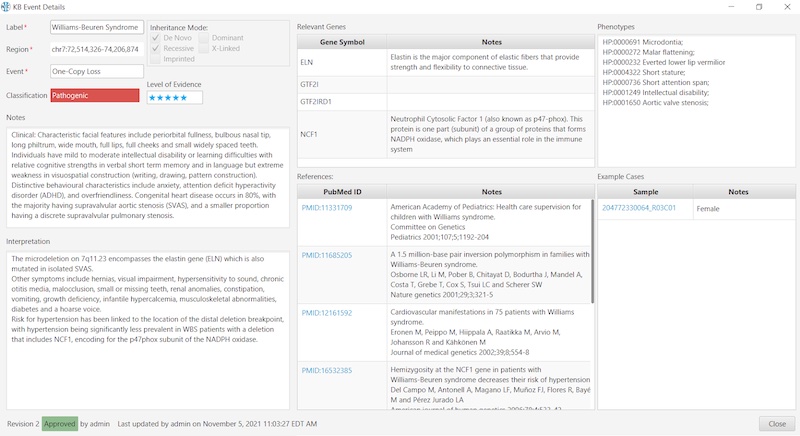
And here’s an example of an Oncology KB entry:
Get the knowledgebase clinical genetics labs rely on to stay on top of demanding, time-sensitive workloads
Here at Biodiscovery, we know labs expect certain things from a knowledgebase. Here are a few of the ways we built this tool for you.
- Ease of use: Making entries in the KB shouldn’t be complicated. In NxClinical, entries are made through a simple, user-friendly form that appears directly in the software interface.
- Dynamic maintenance: Databases aren’t static. They need to be maintained with the most recent information. Our KB is dynamic—allowing labs to make any modifications or updates they need to fit their workflow.
- Auditability: The KB is enabled with a full audit trail to track any submissions and updates within it. (This is a core function of NxClinical.)
“The Knowledgebase we’ve put into NxClinical is unique to this type of commercial software. Clinicians and cytogeneticists have been wanting an easy-to-use database for constitutional and cancer profiles for a long time. Everyone we’ve heard from—NxClinical users and others—has been trying to find bioinformatic ways to gather this information collectively and put it within a few clicks of the user. Our goal with the Knowledgebase is to make NxClinical the gold standard for this.”
— Sam Dougaparsad, PhD, Clinical Market Development Manager, BioDiscovery
New to NxClinical and need an introduction? Let Dr. Brian Lee show you the basics of NxClinical software, the latest features included in the 6.0 release, and its applications to your clinical case review—all in under 30 minutes. Watch our tech tour webinar session →
Considering updating to NxClinical 6.0? In our 25-minute webinar, Making your oncology case review process more efficient through an institutional knowledgebase integrated with NxClinical 6.0, Soheil Shams, Founder & CEO of BioDiscovery, uses multiple example oncology cases to demonstrate the most effective workflow and case review benefits of the Knowledgebase in NxClinical 6.0. Watch the webinar →
Ready to continually enhance your classifications with an integrated genetic knowledgebase that puts your past data to work?
Request a free personalized demo of NxClinical and see the Knowledgebase in action. We’ll connect on an initial consultation to answer questions and dive a little deeper before demonstrating the latest version of NxClinical—either with example data or your own.
*This software is for research use only. It is designed to assist clinicians and it is not intended as a primary diagnostic tool. It is each lab’s responsibility to use the software in accordance with internal policies as well as in compliance with applicable regulations.



