Blog
Aberrant Cell Fraction Estimation in NxClinical 5.0
A little while ago we discussed analysis of CN and LOH in tumor samples which was nicely described in Tumor Profiling: Method and Protocols by Springer. There are challenges in analyzing and interpreting cancer data but the right tools can make this process much easier. The book chapter had noted that mosaicism from a mixture of cells needs to be taken account with cancer samples and that the ability to view allele patterns is important in identifying mosaicism and estimating the % aberrant cells. NxClinical 5.0 has new features to help with this.
NxClinical has very strong visualization tools which is important for cytogeneticists – a reviewer can easily view the allele patterns and estimate % aberrant cells. But NxClinical 5.0 makes this even easier by automatically performing this calculation on a per event basis. The software looks at event-specific aberrant cell fraction using both the Log R value as well as BAF (where available). This approach uses platform-dependent scaling so that the correct calculation can be applied to samples from multiple platforms (e.g. ThermoFisher, Illumina, CNV from NGS). The software also uses a user-defined threshold to mark events as mosaic or not.
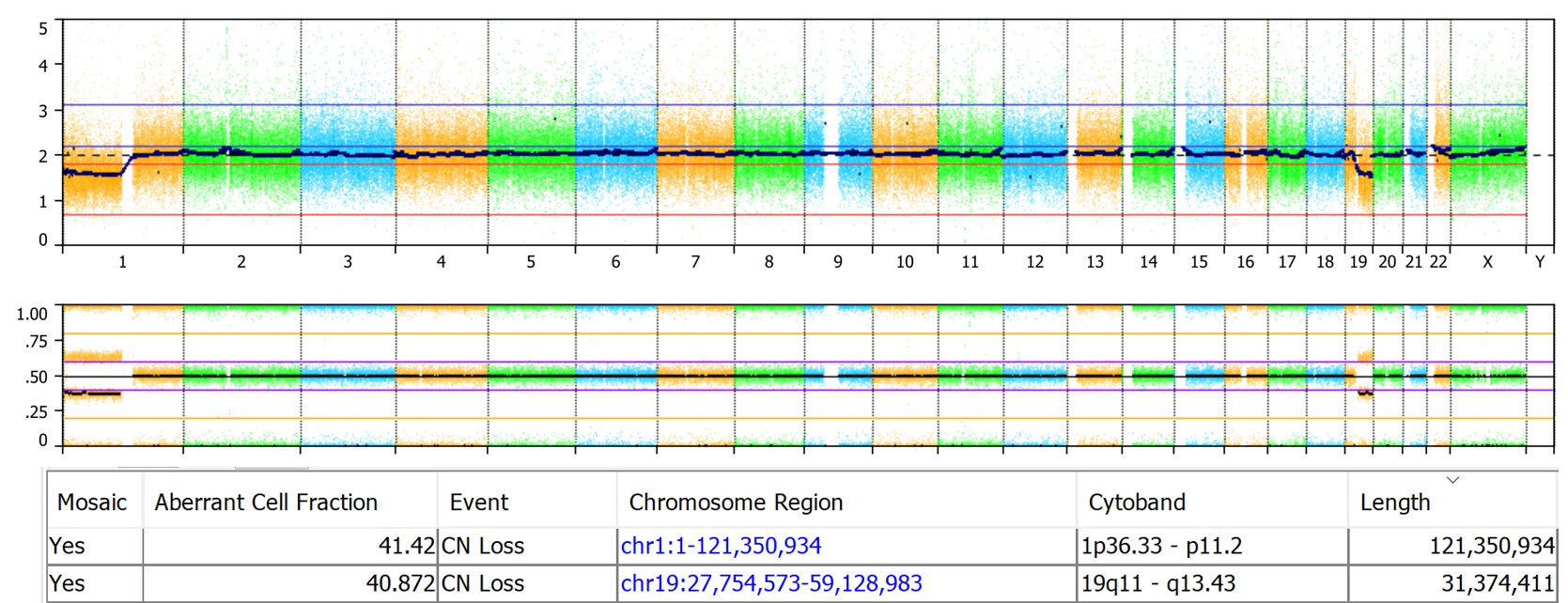
Here’s an OncoScan sample showing mosaic loss of 1p and 19q. The table below the CN probes and BAF plots shows the estimated aberrant cell fraction as 41% for both events. The sample processed via the TuScan algorithm, which estimates a global % tumor value, was identified as having 40% tumor cells which is close to the 41% aberrant cell fraction calculated by NxClinical.
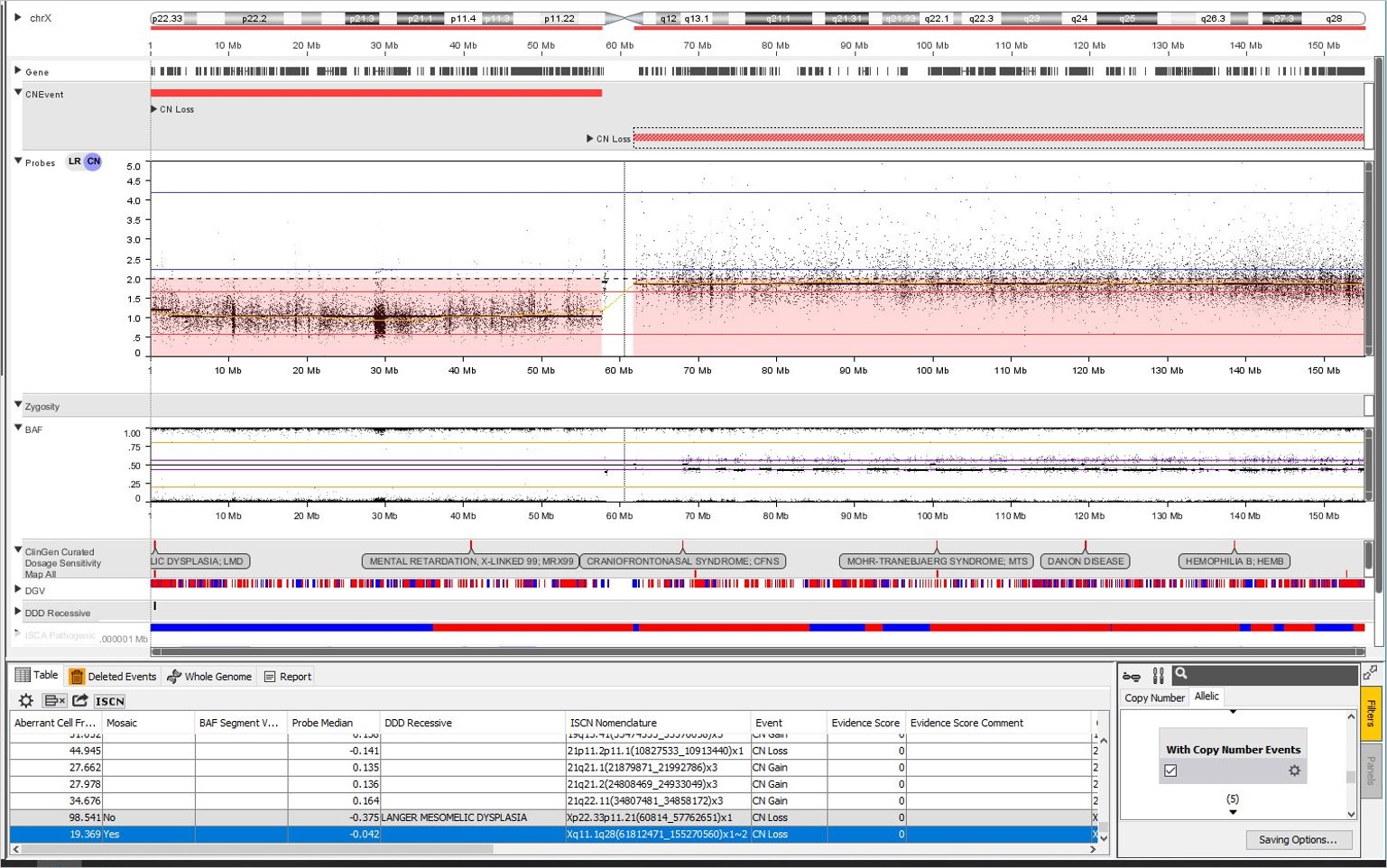
The calculation applies to constitutional samples as well. Here is an Illumina 850K array with a mosaic loss of the q arm of Chr X. NxClinical estimated the aberrant cell fraction as 19%. The mosaicism patterns can be visualized in the CN probes and BAF plots. The CN probes fall below the two copy mark in the CN probes plot and the four band BAF pattern with the middle two bands close to the 0.5 mark indicates low-level mosaicism.
There are limits of detection but overall the software does a good job estimating the aberrant cell fraction on a per event basis. Some platforms have compressed log ratios making it harder to pick up mosaic events; in such cases inspection of the interactive plot visualization allows estimates to be made manually. Typical processing settings for constitutional samples may not be able to pick up mosaic events but this can be overcome by using a set of more lenient cut-offs to pick up mosaic events. The sample can be re-processed with the more lenient settings when mosaicism is suspected and the calculations will be performed automatically.



