Blog
Streamline Analysis of Hematological Malignancies with OGM and VIA™ Software
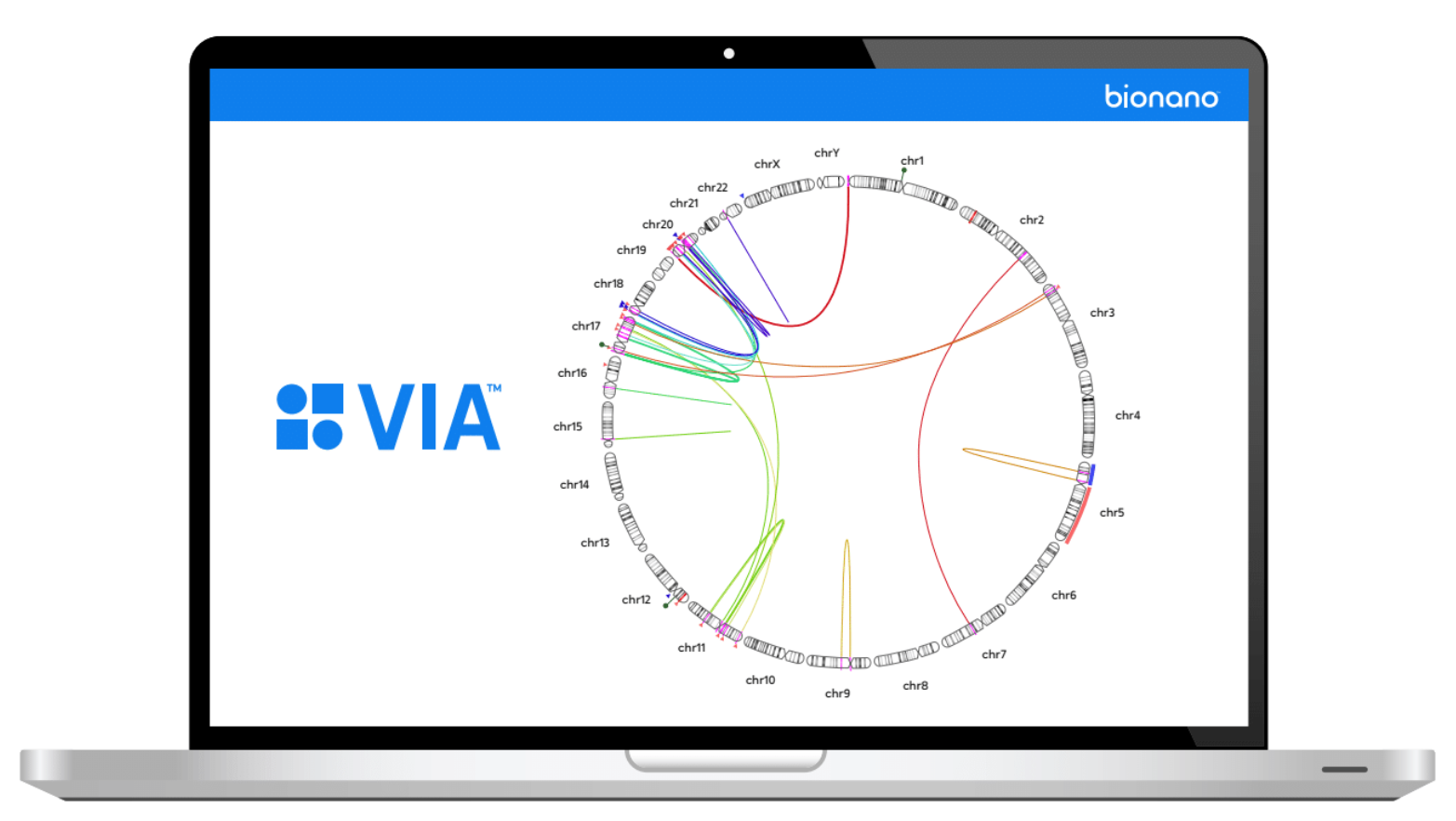
OGM stands as a singular assay capable of detecting all classes of structural and copy-number variants (CNVs) relevant to hematological malignancies, a feat that typically requires multiple cytogenetic techniques. The latest iteration of Bionano software is synchronized to establish a seamless analysis workflow, encompassing primary variant calling, interpretation, and reporting of OGM data. This synchronization is designed to streamline and expedite the identification and reporting of disease-relevant variants.
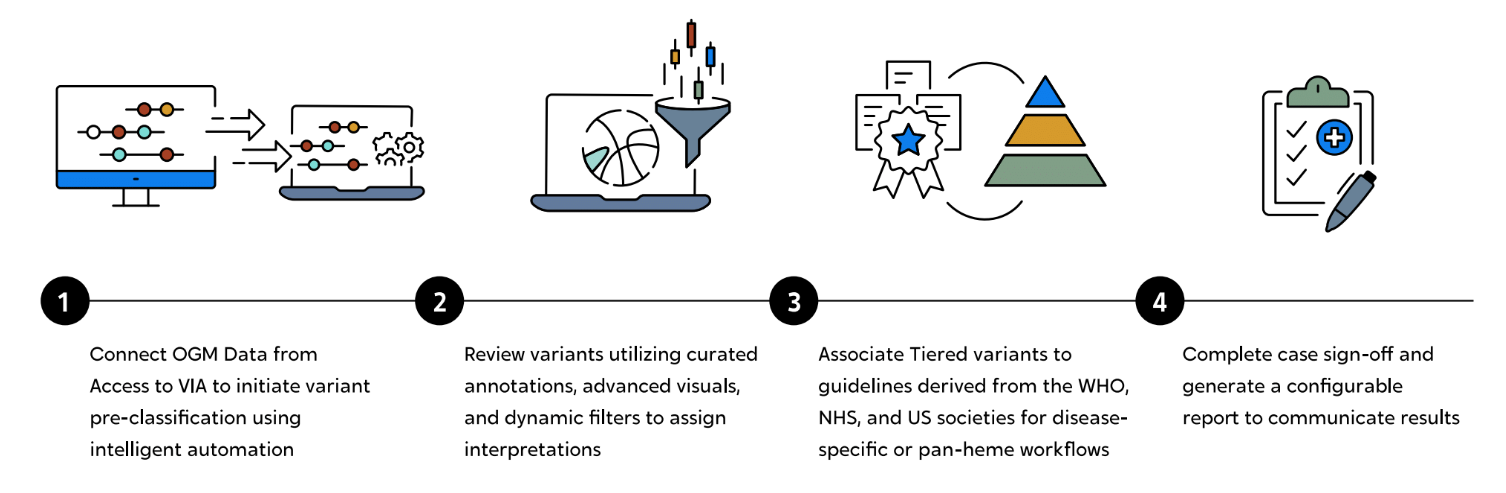
Bionano, through VIA software, integrates curated resources sourced from global professional societies to enhance the analysis of hematological diseases. These society guidelines serve multiple purposes. Initially, they act as targets for automated variant pre-classification, assigning tier status to ensure crucial variants take precedence at the top of the list for analysts. These guideline resources are visually presented in the graphical interface, but also are converted into panels that serve as filters to swiftly focus on key variants of interest. The guidelines, combined with additional genomic annotations, provide relevant information for analysts to interpret and confirm variant tiering. Tier 1 variants can then be associated with a list of the guideline targets to set the detection status for the significant neoplastic events in the sample. To conclude the workflow, a customizable templated report can be efficiently generated, facilitating easy communication of results. The report can encompass essential sample information, variant interpretations, tables displaying society guidelines with detection status, and a summary of whole genome results and data imagery.
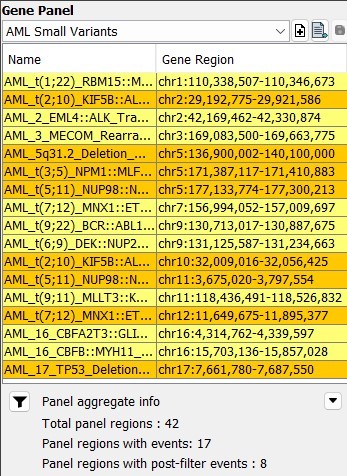
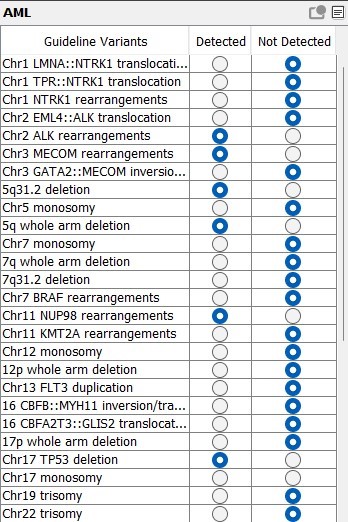
New VIA installations come preconfigured with disease-specific or pan-heme workflows, streamlining the initiation of OGM data analysis for hematological malignancies. However, the software’s full configurability allows adaptation to site-specific preferences. Unveil key structural variants in hematological malignancies effortlessly with optical genome mapping and VIA software, ushering in a new era of precision in genomic analysis.


