Blog
Publication notes superiority of NxClinical software for CNV detection from low pass sequencing
CMA has been the first-tier test for developmental delay/intellectual disabilities, autism spectrum disorders and multiple congenital anomalies but recent advances in next-generation sequencing (NGS) may one day change that standard. Studies have been performed on NGS as an alternative test but costs are of concern particularly with higher depth sequencing (WES, WGS).
A recent publication by Alka Chaubey and team evaluated the potential of Low Pass Genome Sequencing (LP-GS) for detection of CNVs as a replacement to constitutional microarray analysis. The team utilized BioDiscovery’s NxClinical software for CNV detection from the LP-GS test. The authors stated that data analysis for CNV from NGS has been a challenge for clinical laboratories for the last decade. The team evaluated a number of different tools and found NxClinical to have superiority in CNV detection and classification.
“An evaluation of an Edico home brew tool, Variantyx, Golden helix, and BioDiscovery’s NxClinical 5.0 software on the 5X LP-GS data set revealed NxClinical software to have superiority of CNV visualization, analysis, and classification of CNVs for reporting (unpublished data). In addition, challenging samples, such as a mosaic interstitial deletion, mosaic trisomy, and mosaic marker chromosome delineation, were only identified by NxClinical software (Table 1).”
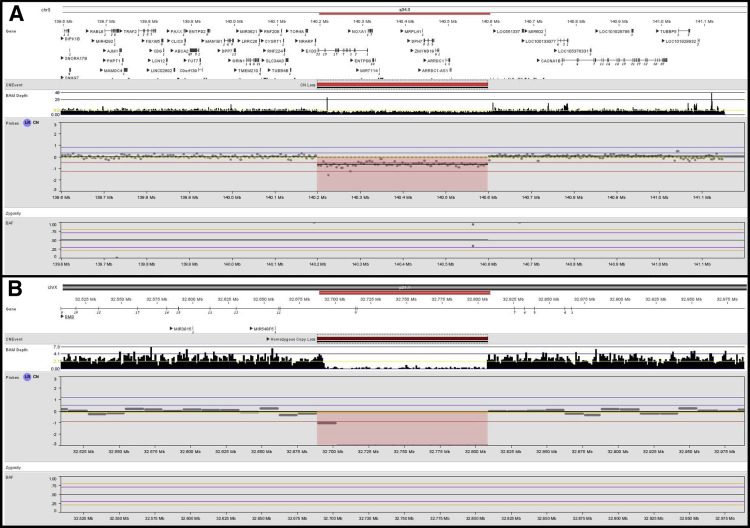
In this study, clinical validation of the LP-GS sequencing method with 5x coverage for sequence variants and copy number events was performed using 78 unique samples that included well-characterized Coriell HapMap samples (NA12878, NA12891, and NA12892). Control samples contained a variety of types and sizes of clinically relevant CNVs and AOH such as trisomies, interstitial and terminal deletions and duplications, mosaic deletions and duplications, absence of heterozygosity, and unbalanced translocations.
The clinical specificity and clinical sensitivity of relevant CNVs was 100% in 33 samples with previously identified CNVs via CMA. In addition, an intragenic single-exon Duchenne muscular dystrophe (DMD exon 55) deletion was detected via LP-GS and NxClinical but had not been detected by CMA due to a lack of exonic probe coverage on the array platform. The test also picked up pathogenic CNVs that were missed by CMA. Seven samples with AOH/LCSH detected via CMA were concordant with the LP-GS test.
Diagnostic yield via LP-GS in a lab setting was evaluated on 331 samples processed at PerkinElmer Genomics Laboratory in Branford, CT. The diagnostic yield of the 5x LP-GS test (17.2%) for pathogenic/likely pathogenic events was comparable to published CMA yield of 5-18% depending on array platform used. In one case of dual diagnosis of Kleefstra syndrome and Duchenne Muscular Dystrophy, the array platform had picked up the EHMT1 intragenic loss but had missed the two exon deletion in DMD. Both these deletions were detected by LP-GS using NxClinical’s MSR algorithm.
A cost analysis revealed a 50% cost savings of 5x LP-GS over microarray which stated by the authors is more cost-effective than any CMA platform being used for clinical testing worldwide. The authors have demonstrated that LP-GS with variant detection and interpretation using NxClinical can be a good alternative in terms of accuracy, diagnostic yield, and cost in replacing CMA for constitutional cases.



