Blog
Come See Bionano’s Groundbreaking DLS Labeling Chemistry at PAG
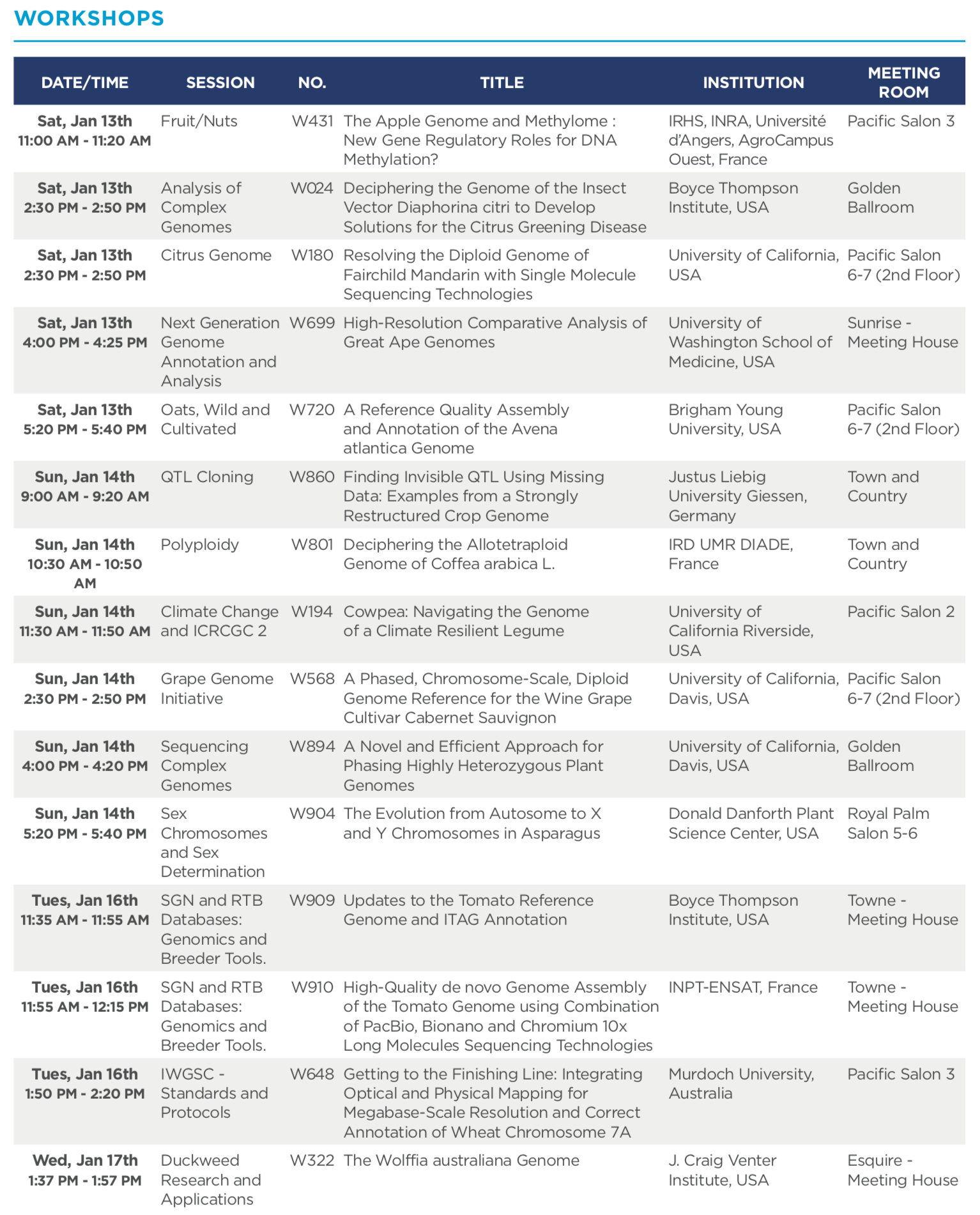
The International Plant & Animal Genome (PAG) conference in San Diego has been Bionano’s favorite for years. After all, the genome assembly community has long embraced Bionano maps to build the best genomes, and the many high profile papers we reported on earlier demonstrate that. At PAG, you can walk into a random session and see Bionano maps on the screen. This year, for edition XXVI, this will be the case more than ever. A total of 23 posters and 16 workshops will discuss genome assemblies and workflows that include Bionano genome mapping data to build the most complete and accurate genome assemblies.
Even more exciting, several presentations will highlight Bionano’s new Direct Label and Stain (DLS) chemistry, to be released later this quarter. At ASHG, we already gave a sneak preview of what this new enzyme and protocol can do. Unlike Bionano’s current Nick, Label, Repair and Stain (NLRS) chemistry, which uses nickases to create a single stranded nick at sequence motifs, DLS uses an enzyme that directly labels the sequence motif. The DLS chemistry does not cause the systematic breaks (fragile sites) that were common with nicking and limited the length of genome maps. Instead, samples labeled with DLS chemistry yield intact maps that are up to chromosome-arm length and capture the linear structure, order and orientation of the genome. During an early access period with over a dozen customers, Bionano’s DLS chemistry has enabled genome maps with contiguities up to 50 times greater than Bionano’s previous labeling method, while maintaining the accuracy of the genome structure Bionano maps have been trusted for. Genomes generated with DLS data have reached scaffold N50s of 100 Mbp, setting a new standard for genome assembly.
Bionano’s genome maps can identify structural variation between individuals, cultivars, breeds or species, and be used to build platinum quality genomes by scaffolding sequence contigs. While other scaffolding technologies improve the contiguity of sequence assemblies by adding additional layers of more complex sequence data, Bionano’s de novo assembled genome maps build a completely sequence-independent, up to chromosome-arm length genome assembly in days for under $1000. Because Bionano’s assemblies are built de novo, without sequence or reference guidance, they can uniquely validate and error correct sequence assemblies while generating genomes of the highest quality and contiguity.
Below you can see all 39 posters and workshops that will be presented, which include an oral presentation on DLS chemistry by a Bionano scientist (W025), and presentations by DLS early access customers from the Vertebrate Genome Project lab at Rockefeller University (P0204), and Genoscope (P0176). The Bionano presentation will feature assembly data generated with DLS on a dozen organisms such as fish, birds, wheat, maize, Brassica, mammals and human.
This year we are hosting a “Meet the Experts” panel at Bionano booth #523 where a few early access DLS users will be available to discuss the impact the new chemistry is having on their assembly projects.
Meet the Experts
Date: Monday, Jan 15th, 2018
Time: 11:00 AM – 12:00 PM
Location: Bionano Booth #523
With the upcoming launch of our Direct Labeling chemistry, Bionano will set a new standard for genome assembly quality. The record number of presentations that include Bionano data at PAG shows that Bionano mapping is an essential tool for genome researchers. With DLS, we will greatly increase the accuracy and contiguity of plant and animal genomes, and 2018 promises to be an exciting year for Bionano and for the genome assembly community as a whole!
Come see us at booth #523 at the PAG XXVI. Download the brochure with all Bionano presentations here.
Bionano Genomics PAG XXVI Oral Presentation
W025: Building Maize High Quality, Chromosome-Scale, de novo Genome Assemblies by Scaffolding Next-Generation Sequencing Assemblies with BioNano Maps Generated with the New Direct Labeling Enzyme
Speaker: Yang Zhang, Bionano Genomics
Saturday, Jan 13, 2:50 PM -3:10 PM
Session: Analysis of Complex Genomes
Location: Golden Ballroom