Blog
How to Identify Chromothripsis in Tumor Samples
Chromothripsis, or chromosomal shattering, occurs at a frequency of 1%-5% in most cancer types, though has been described as being upwards of 25% in bone cancer (Cell. 2011 Jan 7;144(1):9-10). It has been associated with poor prognosis (Cancer Res. 2012 Dec 27). Chromothripsis has even been found to occur in the germline of some developmental disorders (Hum Mol Genet. 2011 May 15;20(10):1916-24, Cell Rep. 2012 Jun 28;1(6):648-55.). This complex chromosomal rearrangement is thought to occur when the chromosome breaks apart into tens or even hundreds of pieces and is then rejoined through nonhomologous end-joining. While a primary indicator of chromothripsis is a highly rearranged sequence, several hallmark features of chromothripsis can be observed in BioDiscovery Nexus Copy Number software, including alternating copy number states, high level of breakpoints, and loss of heterozygosity in the lower state.
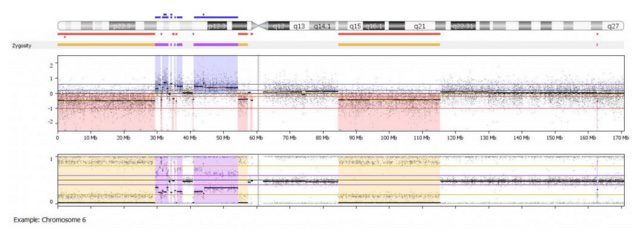
In the example shown above, we can clearly see several regions of copy number loss (red regions in middle logR panel), which are associated with appropriate loss of heterozygosity (yellow regions in the bottom B-allele frequency panel). These regions alternate with copy number neutral regions (uncolored baseline in logR panel, uncolored 3 band pattern in bottom B-allele frequency panel) and regions of copy gain (blue regions in logR panel, purple 4 band allelic imbalance pattern in B-allele frequency panel).
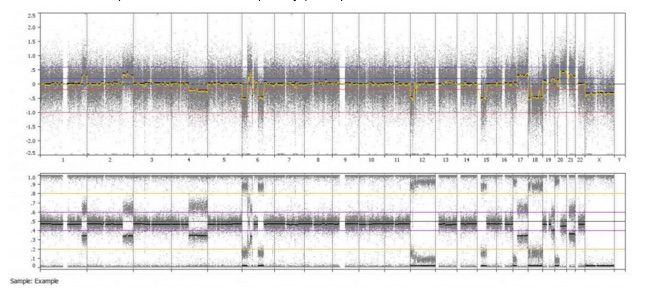
Even from a whole genome overview, we can easily identify the alternating copy states and altered heterozygosity of chromosome 6. This is just one of the many ways BioDiscovery Nexus Copy Number software can be used to identify some of the frequent aberrations found in cancer.
You may also like to read the article here on how to estimate the percentage of aberrant cells in a tumor/normal cell mixture.
