Blog
Elevate Your Genomic Insights with OGM and VIA™ Software

OGM isn’t just a tool; it’s a breakthrough, detecting all structural and copy-number variants crucial to hematological malignancies in a single powerful assay. Seamlessly synchronized with the cutting-edge Bionano software, VIA revolutionizes your analysis workflow, accelerating the identification and reporting of disease-relevant variants. Combining OGM and VIA software delivers a comprehensive and automated workflow for analyzing, interpreting, and reporting structural variants (SVs) and copy number variants (CNVs) associated with hematological malignancies.
This complete workflow leverages intelligent software automation, reducing the time to result for hematological malignancy samples from OGM data. VIA software includes curated resources representing guideline-based targets applicable to hematological disease, pre-classifying variants detected with OGM according to a rules-based engine for efficient interpretation of pathogenicity.
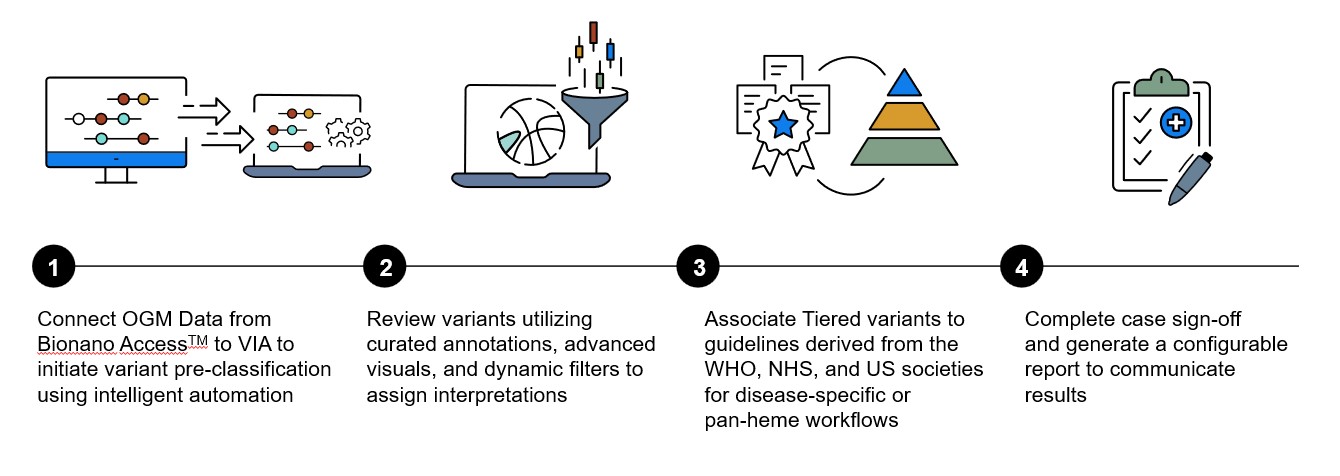
Experience the enhanced capabilities of VIA 7.1 software, offering additional features and usability enhancements to further optimize the analysis workflow for hematological malignancies. New functionality has been developed to improve visualization, interpretation, and reporting of OGM data, providing a comprehensive assessment of hematological diseases.
Advancements in Structural Variant Detection and Quality
OGM with VIA software enables the detection of all classes of structural variants impacting hematological disease with high sensitivity and precision. VIA 7.1 includes a new feature to confidently call aneusomies as low as 5% VAF for inclusion in the sample report. Read more about the new aneusomy caller.
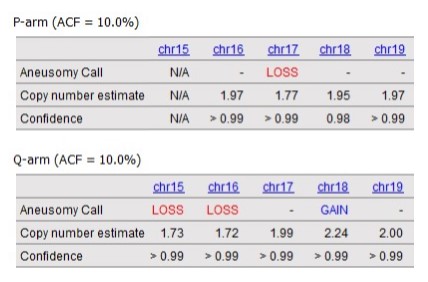
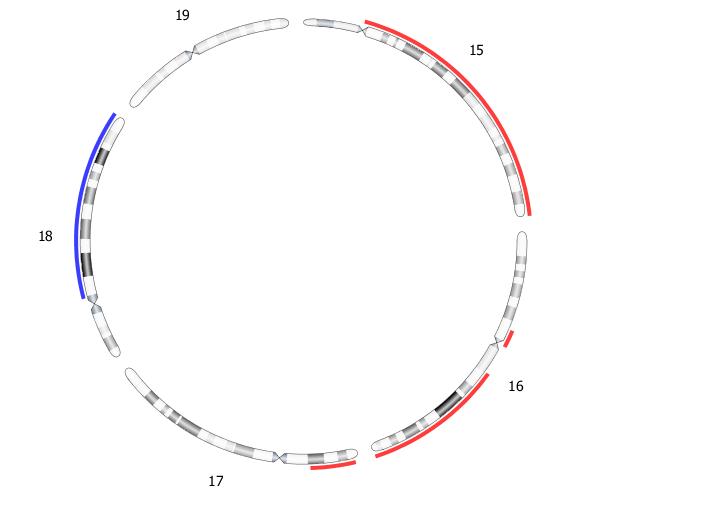
Example aneusomy table results with corresponding Circos plot
Bionano Solve provides powerful informatics pipelines to detect novel structural variants from OGM data. The newest pipeline for structural variant calling, Guided Assembly LAF, employs a reference-guided approach for whole genome assembly, increasing sensitivity for detecting small structural variants at low allele fractions while enhancing precision for negative results. New in VIA software, detected SVs are assigned quality scores on a phred scale to standardize identification and filtering of high-confidence SV calls. Structural variants with an SV quality score of Q20 signify that the variant was confidently called with over 99% certainty. Standardizing the quality scoring of SVs heralds a new era of efficiency and confidence in identifying disease-relevant SVs for interpretation.
Empowering Insightful Visualization with the Dynamic VIA Circos Plot
Visualization is key in cytogenetic analysis, providing critical context for variant interpretation. VIA software generates powerful visuals of relevant datasets, consolidating critical information into a single view to interpret OGM data. Interacting with structural variant data through a Circos plot provides analysts with a clear perspective on how multiple SVs could impact the genome. The Circos plot in VIA 7.1 has numerous enhancements to facilitate yielding new insights from your data.
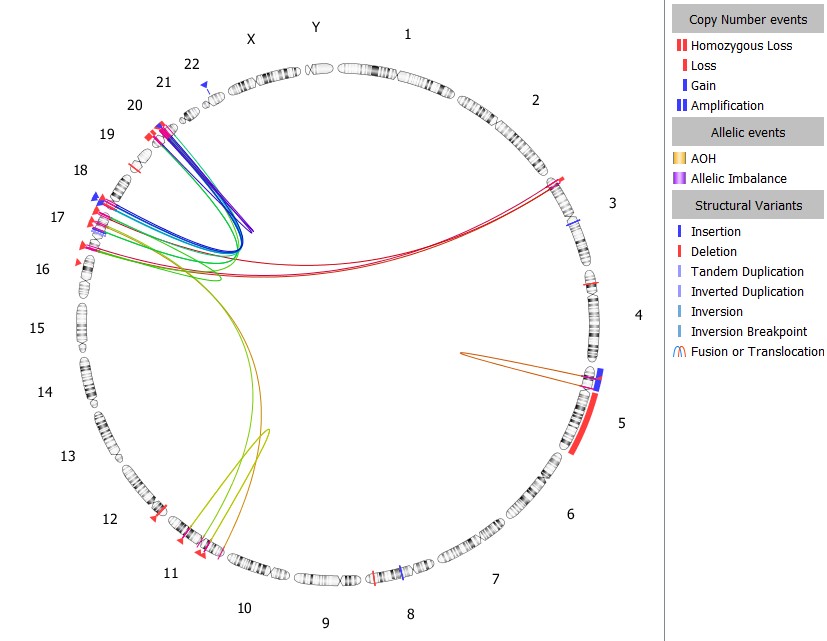
The VIA Circos plot offers a comprehensive visual representation of all events that have passed through the variant filters, presenting a consolidated view of SVs relevant to the sample analysis. Its dynamic display allows for the simultaneous visualization of both intra-chromosomal and inter-chromosomal events. With VIA 7.1, the Circos plot offers enhanced clarity and context by enabling users to dynamically magnify chromosomal regions with complex events.
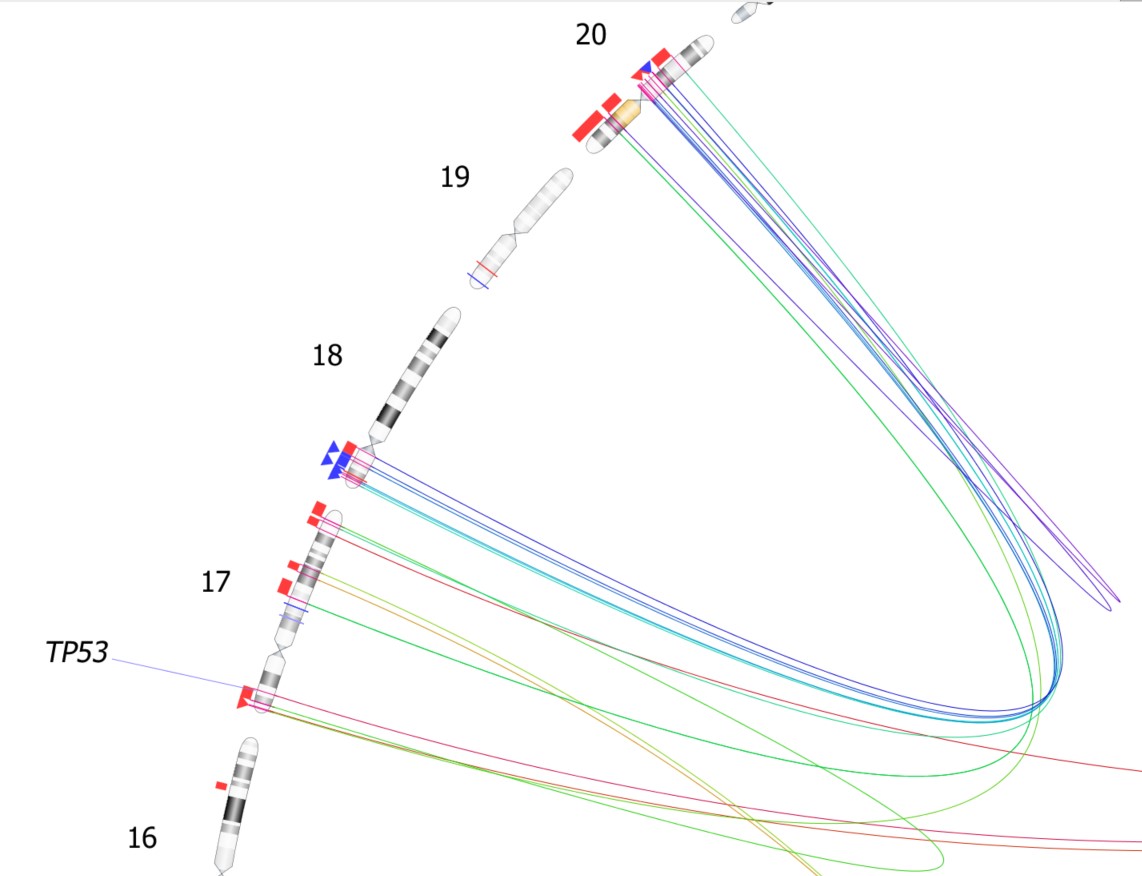
Moreover, VIA 7.1 allows for the inclusion of key genes of interest alongside the chromosome position of SV occurrences, emphasizing the significance of variants to the investigated condition. Annotating the Circos plot with genes is made simple with VIA’s user-friendly interface, facilitating clear emphasis on impacted genes. Additionally, the Circos plot in VIA enables users to selectively focus on specific chromosomes, streamlining the display of SVs affecting those chromosomes for a concise visual representation when necessary.
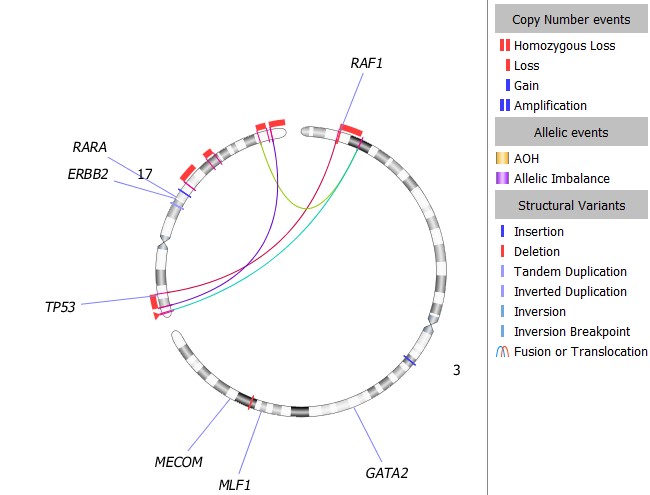
Enhancing Interpretation Efficiency and Accuracy
OGM provides a single assay to detect chromosomal aberrations of biological significance at higher resolution compared to classical cytogenetic techniques. VIA software aids interpretation using sophisticated approaches to automatically classify variants, filter against curated datasets, and remove irrelevant variants. VIA 7.1 introduces enhancements to automated structural variant classification, utilizing critical SV data such as quality, frequency, and size to accurately identify disease-relevant SVs with optimal performance.
Effortless Reporting with Enhanced Data Visualization with VIA Software
Easily create reports with VIA’s customizable reporting options, communicating results with simplicity and clarity. Bring the most relevant information to the surface along with visuals and data that support decision-making that meet guideline-based requirements. In VIA 7.1, important imagery, including Circos plots, Whole-Genome plots, and Ideograms, can be automatically included in reports to ensure visual representation of data. Share insights derived from OGM data in VIA software with key stakeholders effortlessly.
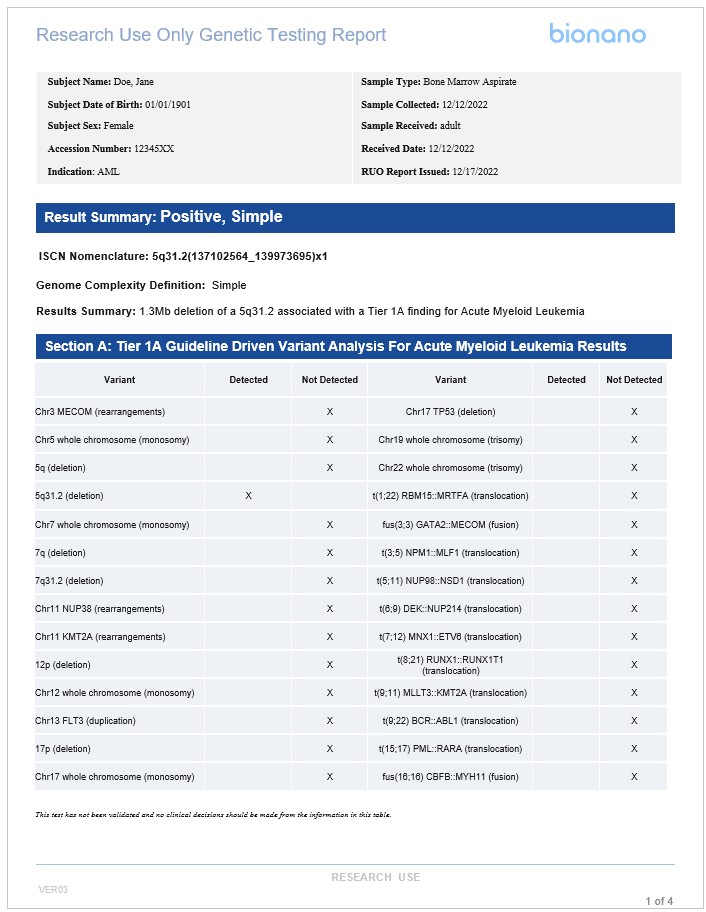
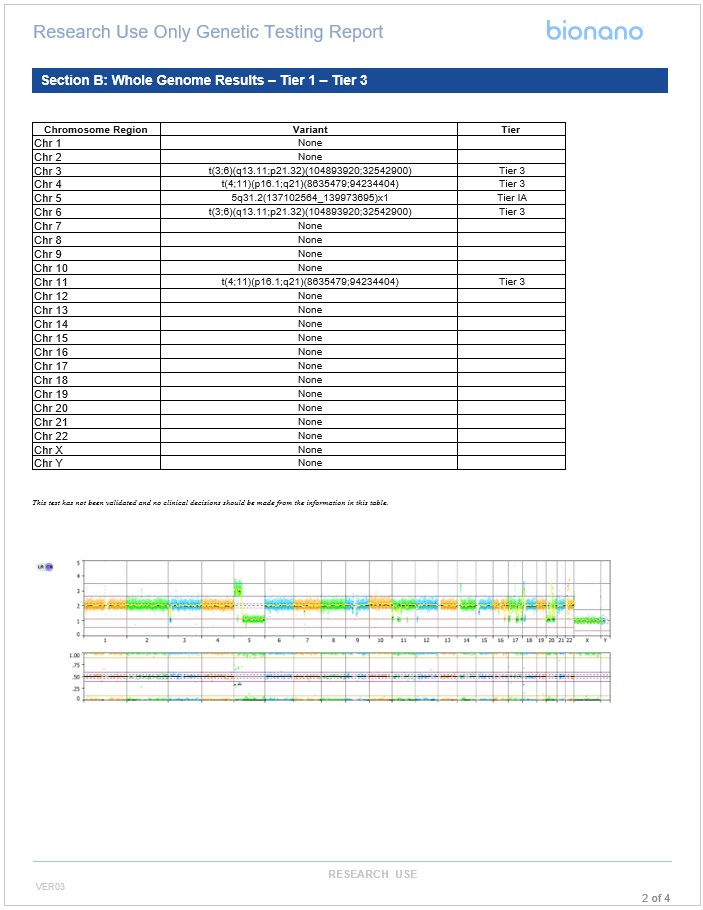
Example templated report for hematological malignancies
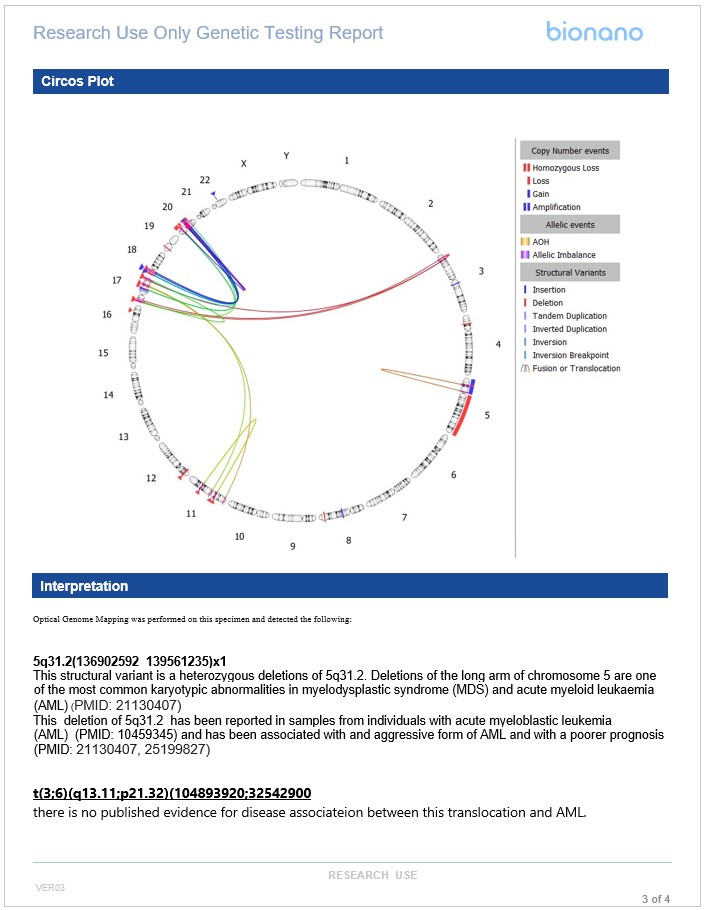

Example templated report for hematological malignancies
Brian Lee, PhD, Software Application and Customer Solutions Manager at Bionano, dives into software architecture and OGM sample analysis features in VIA.
Talk to a specialist to discuss how you can benefit from VIA software in more detail or request a demo to review the software’s capabilities in detecting and analyzing genome-wide variation associated with hematological malignancies.