Blog
The DGV score and so much more
For almost two decades the Database of Genomic Variants (DGV) has provided researchers and clinicians open access to common copy number variations, from diverse genomic populations for the purpose of identifying common polymorphisms in these populations. Early DGV data was generated mostly from low-resolution microarrays and the quality of the data was questionable in terms of samples being truly “normal”. More recently, data from massively parallel next-generation sequencing (NGS) and much better-controlled sources have been added to DGV including CNV data from the Gnomad database1.
What is DGV Gold?
In 2019, the DGV group published a paper and released a new sub-track called DGV Gold Standard which is a curated dataset of variants2. This was created by applying quality filters and merging overlapping variants to the existing DGV dataset in an effort to remove low-resolution data and reduce the number of polymorphic variants3. At the time, the existing DGV database held variants from 75 peer-reviewed studies whereas the new DGV Gold captured variants from only 32 of those studies because at a minimum a variant must be identified in at least two different studies and located in at least two different samples. This new sub-track allowed for increased sensitivity and increased specificity as it clustered variants with a minimum 50% overlap and removed low-resolution studies with high false-positive rates, respectively4.
Every year, more and more published structural variants are being reported and yet the DGV Gold Standard Dataset has not been updated since its original release in the summer of 2019.
What is the latest DGV Track in NxClinical?
The DGV track in NxClinical is not an exact copy of the data available on the DGV website. BioDiscovery curates this data to provide a higher quality set of variants before presenting it in NxClinical. As of the Spring 2021 track updates, the data in the NxClinical DGV track starts with all available studies (including CNV data from Gnomad) and is then filtered based on the following criteria:
- Remove all studies that included BAC results (as these are not of high quality and very old)
- Remove all events less than 50bp (to filter out seq var changes from sequencing data mostly reported by GnomAD-SV)
The resulting track then contains more studies (including newer ones) than the DGV Gold and does not include any low-quality data.
What is DGV Score and how can I use it?
The main purpose of using the data in the DGV is to see if an observed CNV in the sample under review is likely to be benign due to its observation in an unaffected population. To make that determination, there are two main factors to consider. First, how similar is the event in the sample to those reported in the DGV, and second, how many times has that event been seen in the general population?
The DGV Score calculated in NxClinical combines these two factors into a single score ranging from zero to one. This combined score is calculated by considering all studies that report a CNV event that overlaps the event of interest. It then looks at the similarity between the event and the one reported in DGV using the Jaccard Index. This score is calculated by dividing the length of overlap between the two regions by the total length (union) of the regions as depicted in the image below.
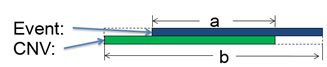
Once the similarity is measured, it is scaled using a non-linear transform where a single reported observation scales the similarity by 60% and observation of more than 30 times scale by 98%. So, if the event overlaps a region in DGV perfectly but it has been reported a single time, the DGV score will be 0.6. Alternatively, if an event overlaps a DGV entry that has been reported 100 times, but with a similarity of 0.6, it will still have the same score of 0.6. This approach ensures that to get a score of greater than say 0.9, both the similarity has to be high (e.g., greater than 0.9) and reported many times (possibly more than 20 times).
Where do I see the DGV Score during case review within NxClinical?
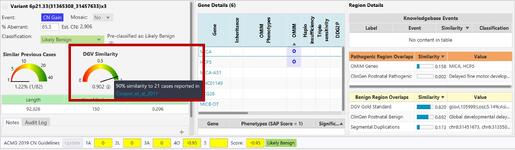
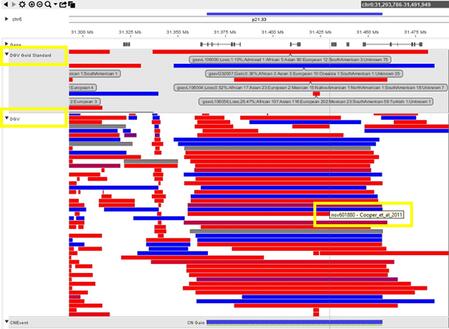
How can BioDiscovery’s curated DGV track help you?
Offering the DGV track in NxClinical is a necessity but BioDiscovery takes it to the next level by curating the database to a gold-standard offering where you can have peace of mind knowing the variants you’re viewing have been added by the latest technologies and aren’t composed of outdated findings.
If you think NxClinical may be the right variant interpretation software for you, learn more from this 30-minute tutorial “What is NxClinical and how can it help my clinical case review?”
References:
1. MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014 Jan;42(Database issue):D986-92. doi: 10.1093/nar/gkt958. Epub 2013 Oct 29. PMID: 24174537; PMCID: PMC3965079.
2. Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, Dougherty ML, Nelson BJ, Shah A, Dutcher SK, Warren WC, Magrini V, McGrath SD, Li YI, Wilson RK, Eichler EE. Characterizing the Major Structural Variant Alleles of the Human Genome. Cell. 2019 Jan 24;176(3):663-675.e19. doi: 10.1016/j.cell.2018.12.019. Epub 2019 Jan 17. PMID: 30661756; PMCID: PMC6438697.
4. DGV Struct Var Track Settings
Additional resources:
DGV (Database of Genomic Variants [*** v107 ***] )
CNV Technical Standards Web Series – ClinGen | Clinical Genome Resource



