Blog
Copy Number and Loss of Heterozygosity Analysis in Tumors
If you are new to cancer analysis and need some direction on CN and LOH analysis in tumors, take a look at Tumor Profiling: Methods and Protocols published by Springer. The 2019 book is out and Chapter 7 “Whole-Genome Single Nucleotide Polymorphism Microarray for Copy Number and Loss of Heterozygosity Analysis” by authors Ross Rowsey, Iya Znoyko, and Daynna Wolff is an excellent resource for analysis of tumor samples via SNP microarray. The chapter begins with acknowledgement that copy number (CN) changes are more prevalent in many cancers than are single nucleotide mutations and the optimal method to detect such somatic changes is via SNP microarrays. A nice summary of the SNP microarray platform and with a focus on data analysis is presented.
The authors highlight advantages as well as drawbacks of SNP microarray as compared to karyotyping and sequencing technologies. For example, SNP microarray can handle DNA extracted from many sources directly (not requiring cell culture) including liquid tumors, fresh tissues, and even FFPE which cannot be used in conventional karyotyping. Single gene deletions involving tumor suppressors or focal amplifications of activating oncogenes may not be easily identifiable via sequencing but can be by microarray analysis.
Copy number state is determined by probe intensities in log 2 scale and the chapter explains the basics of how this is accomplished (there are variations among different algorithms) along with a nice summary of allelic event display using B-allele frequency data obtained by SNP arrays. An important point made in this guide is that thorough interpretation requires knowledge of the various patterns created by CN and allelic changes as a combination of these allows for ascertainment of all valid calls in the array data. Visualization tools are extremely important in this process as the patterns in log ratio and BAF plots need to be reviewed by an individual to make the calls. The authors show examples of these patterns with some of the figures (7 and 9) generated via NxClinical software (for analysis and interpretation of CNV, allelic events, and sequence variants). Visualization is one of the strongest features of NxClinical software; it plots the underlying raw data (probe and BAF) used to make gain, loss, and LOH calls in an interactive browser along with reference data to be used for interpretation.
A major challenge with cancer samples is that they can have different ploidies. The authors note that many microarray analysis software tools are designed for analysis of constitutional samples and assume that the copy number state of the genome is two providing an example of triploidy (69,XXX) where the genomic copy state appears normal using LogR ratio as everything is normalized to a copy number of two. The true ploidy can only be unveiled by examining the SNP probes where four allele clusters are present for all chromosomes. It’s crucial for the reviewer to be aware of the patterns created and dig down into the probe profiles. Examination of the BAF along with the log ratio can help determine the baseline ploidy. In figure 9, the authors show that correcting the ploidy of a renal cell carcinoma (RCC) sample transforms the results into a CN and LOH profile resembling the classical pattern observed in chromophobe RCC. The algorithms in NxClinical use BAF data with the Log R to make more accurate gain/loss calls as well as ascertaining LOH and allelic imbalance. The ability to view the underlying probe data to discover these “patterns” and re-center to correct for ploidy in NxClinical is very helpful to cancer cytogeneticists.
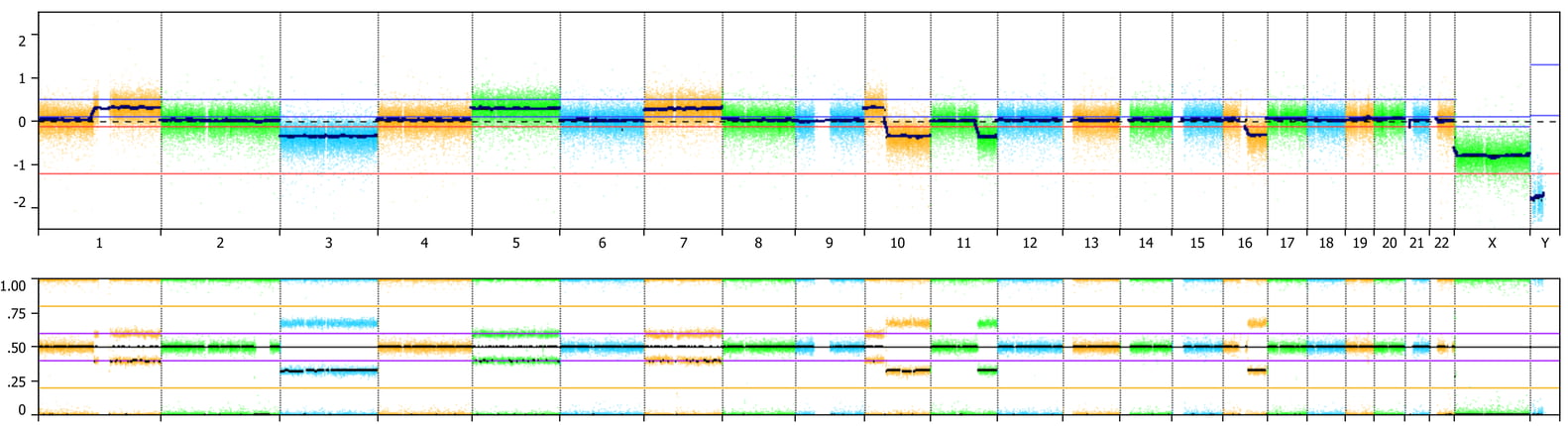
Another aspect of cancer samples to take into consideration is mosaicism resulting from a mixture of cells (e.g. mix of tumor/normal cells in the specimen or multiple clones in the tumor). The authors note that allele patterns are important in identifying and estimating the percentage of mosaicism. Some tools such as NxClinical estimating this value and automatically annotate events as mosaic or not making the review process much faster and easier. There are some new features in the upcoming NxClinical 5.0, including % aberrant cell fraction, that help with cancer analysis and I’ll provide an overview of this in the next post.
The chapter on CN and LOH analysis and interpretation in tumor samples in this book provides a lot of great information and guidelines on analysis and interpretation of tumor samples. The authors focus on the data analysis portion and emphasize the importance of visualizing the underlying raw data to assist with interpretation. I highly recommend getting your hands on this book and reading this chapter.


