Blog
Automating Genomic Scar Analysis For HRD
If your time is short:
- Homologous recombination deficiency (HRD) is a cell’s inability to repair DNA strands resulting in an acquired rate of chromosome breaks.
- HRD scoring is an analytical approach to measuring the cellular rate of acquiring chromosome breaks in specific, quantifiable patterns, or “genomic scars.”
- Cells with elevated HRD scores have been shown to be more sensitive to poly (ADP-ribose) polymerase (PARP) inhibitors and platinum-based therapies.
- Single nucleotide polymorphism (SNP) arrays and Next-Generation Sequencing (NGS) are both commonly used methodologies for detecting HRD. Both methods are designed to detect copy number variants (CNVs) and loss of heterozygosity (LOH).
- NxClinical™ software, built by BioDiscovery, a Bionano Genomics company, gives clinical research labs an analysis solution to conveniently and reliably detect genomic scars that enables the calculation of HRD scores from SNP arrays or NGS data.
Learn more about NxClinical software and request a demo on our website to see it in action.
Homologous recombination deficiency (HRD) represents a major area of research that is focused on demonstrating the utility and characterization of genomic scar analysis for implementation into routine care settings.
Through their involvement in the Friends of Cancer Research (FOCR) HRD Harmonization Project, partner labs are using automated genomic scaring analysis in Bionano’s NxClinical software. This tool will increase their ability to perform the genome-wide data analysis needed to measure and assess the performance of current and future HRD assays. FOCR collaborators will then use this data analysis to coordinate the use of HRD as a biomarker to guide certain treatment types.
Here, we briefly explore the background and current understandings of HRD, its potential clinical research applications, and how the latest version of our genomic analysis software, NxClinical 6.2, provides powerful new capabilities for research labs looking to calculate HRD scores.
What is HRD?
HRD is the inability to repair DNA double-strand breaks using the Homologous Recombination Repair (HRR) pathway.
This repair deficiency results in increased accumulation of chromosomal structural variants in the form of deletions, duplications, loss of heterozygosity (LOH), translocations, insertions, and inversions.
HRD is a common hallmark of cancer, making it of high clinical interest due to the sensitivity of homologous recombination-deficient cells to poly (ADP-ribose) polymerase (PARP) inhibitors and platinum-based therapies.1
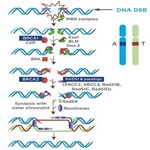
Overview of HR Repair
(Graphic adapted from Iliakis et al2)
Stover, et al offers a more detailed definition of HRD:
“Homologous recombination DNA repair deficiency (HRD) is a functional defect in homologous recombination DNA repair, arising from germline or somatic mutations in BRCA1/2 or other mechanisms. Cells with HRD are more sensitive to platinum and poly (ADP-ribose) polymerase inhibitors (PARPi). HRD generates permanent changes in the genome with specific, quantifiable patterns (‘genomic scars’).”
Harmonizing HR Status and its Use as a Biomarker in Clinical Research Settings
Homologous recombination repair is one of the major mechanisms of defective DNA repair and frequently occurs in cancer. It’s emerging as both a promising biomarker and treatment vector for some disease types.
The genomic scarring often left by homologous recombination is now the subject of intense research and discussion to understand and reach a consensus on what information HRD contains pertaining to a current or potential disease state.
Right now, however, there is no standardized way to define, measure, and report HR status—a major roadblock to understanding and potentially realizing its clinical utility.
The Friends of Cancer Research, in partnership with a large working group of which Bionano is a member, is currently working on the HRD Harmonization Project to solve that problem. This project aims to understand the differences in the current assays available today for HRD and ultimately aligning methods for measuring HR status and its use as a clinical biomarker.
Briefly, the project timeline has been divided into three phases:
- Phase 1 has been completed and involved analyzing the landscape to understand current practices in the field regarding HR status. Results of that analysis can be found in the publication Homologous Recombination Deficiency: Concepts, Definitions, and Assays.3
- Phase 2 involves measuring variability by comparing results from multiple assays that measure HR status. More than ten diagnostics developers will work together to create an analysis plan. These developers will run their HR assays on two different ovarian cancer datasets (The Cancer Genome Atlas tumor samples and archived patient tumor samples), and then the group will compare results to describe concordance and propose potential sources of discordance.
- Phase 3 involves proposing harmonized approaches for generating meaningful clinical evidence. Friends will convene stakeholders to develop an approach to disseminate findings and contextualize the clinical value of HR status.
Through deployment of our genomic data analysis solution, NxClinical software, we are able to contribute an analysis modality that allows for the detection of genomic scars associated with HRD.
The Clinical Research Application of HRD Analysis and Scoring
Recent work in germline and tumor sequencing has provided a better understanding of HRD and its potential role as a genetic contributor to certain diseases, particularly the development of some cancers.
Today, evidence suggests that HRD impairs normal DNA damage repair. This, in turn, can result in loss or duplication of chromosomal regions or genomic loss of heterozygosity. As we mentioned earlier, cancer cells with these functional defects, expressed as elevated HRD scores, have been shown to be more sensitive to certain routes of treatment, namely poly (ADP-ribose) polymerase (PARP) inhibitors and platinum-based therapies.
Detecting the presence of and quantifying the degree to which HRD is present in a solid cancer tumor may provide immense clinical utility in making optimal treatment decisions.
The Challenges of HRD Measurement and Scoring
Measurement of HR status employs two key approaches: a causative assessment of point mutations within genes associated with the homologous recombination repair cellular pathway, and an assessment of the functional loss of HRR mechanism. A functional assessment of the tumor’s genomic instability has previously been challenging due to the limitation of technologies to confidently assess structural changes of the tumor. A few methods for detecting and measuring HRD have emerged over time.
- Some methods involve the assessment of sequence variants, particularly germline or somatic pathogenic variants (PVs) in HR pathway genes (primarily BRCA1/BRCA2) that have been well documented in clinical research studies.
- Other methods assess the genomic instability that results from HRD by looking at three specific genomic measurements of DNA repair competency: loss of heterozygosity (LOH), telomere allelic imbalance (TAI), and large-scale state transitions (LST).

Two analysis techniques are typically employed to use these methodologies: SNP arrays and NGS.
Right now, HRD analysis and scoring is not a common assessment capability in most clinical research labs. Simply put, it’s too specialized for these labs to justify the investments necessary to add the required technology to their toolkits. In addition to the software requirements, most labs often do
not have access to the data required for the software to perform an analysis, further prohibiting scoring capabilities.
Should a better understanding of the utility of HRD analysis develop, many clinical labs may find themselves without the technical capabilities to efficiently assess the consequences of HR deficiency. (More on how we’re solving this problem in the next section.)
Another limitation of many labs measuring and scoring HRD today is that these labs are often only using LOH analysis, rather than a combination of LOH, TAI, and LST, which has been demonstrated to have a higher value than the LOH score alone.4
Solid tumor tissue storage layers on additional technical challenges that can further complicate the prospect of HRD analysis. This limitation needs to be considered when including genomic scar analysis into a workflow. However, NxClinical software is capable of conducting a genomic scar analysis from methods adapted for FFPE tissues.
Genomic Scar Analysis in NxClinical Software

The most recent version of our software takes these capabilities to the next level—automating formerly manual scoring and displaying those numerical values in a convenient view.
NxClinical Software’s Genomic Scar Analysis and Scoring Workflow
Here’s a brief primer on the NxClinical software’s genomic scar analysis workflow:
- Detect copy number (CN) and LOH events from arrays and NGS. This involves merging CN and b-allele frequency (BAF) event tracks (performed once and used for all three scores: LOH, TAI, LST).

- Merge and smooth similar event types for scarring. Once NxClinical software has a unified representation—one track—it smooths the resulting merged track to combine similar event types, omit small gaps, and join events spanning the centromere.
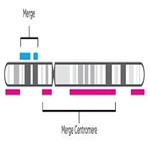
- Select the resulting events that meet each scar’s criteria (LOH, TAI, and LST).
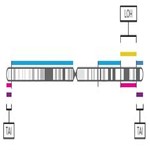
- Visualize the genomic scars genome-wide.

Karyogram visualization of the copy number regions impacted by genomic scaring instability.
HRD Genomic Scar Definitions in NxClinical Software
- Loss of heterozygosity (HRD-LOH)
The number regions of representing one parental allele via LOH longer than 15 Mb but shorter than the whole chromosome5 - Telomeric Allelic Imbalance (TAI)
The number of regions of contiguous allelic imbalance that extend to one of the subtelomeres but do not cross the centromere6 - Large-Scale State Transition (LST)
The number of chromosomal arm breakpoints between adjacent regions greater than 10MB but not separate by less than 3MB7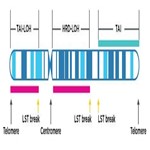
Schematic demonstrating example measurement of a chromosome leveraging a comprehensive genomic scarring approach. It is possible for events to meet the criteria of multiple scars while other events will not count towards the scores.
What’s New in NxClinical 6.2
The latest version of NxClinical 6.2 standardizes the analysis approach we just described and conveniently integrates the resulting genomic scar measurements in user reporting.
This approach gives labs a more scalable means to analyze genomic scars, reducing steps—and time—from the workflow without compromising the reliability of analysis.
- All the additional steps of merging, smoothing, and adjoining are run automatically via more sophisticated logic that protects against overcounts and undercounts. NxClinical 6.2 essentially takes the chromosome together as a whole, as opposed to just looking at individual events, which could risk double counts.
- New breakpoint capabilities also offer additional value. With NxClinical 6.2, labs can conveniently assess the genome to identify these state changes and display them in context.
See More of The Tumor Profile
Perhaps the strongest benefit to NxClinical software for HRD analysis and scoring is the enhanced transparency and clarity of chromosomal abnormalities, which enables teams to analyze HRD more accurately.
With NxClinical software, labs can look at a sample in its entirety, can better understand the impact allelic copy number has on chromosomes and genomic scarring, and can better visualize the impact of HRD, such as seeing the deletion of a critical oncogene. Perhaps it’s not just the gene that’s deleted, but rather the entire chromosome arm enabling more insights for clinicians and supporting further understanding.
In short, with NxClinical 6.2, labs are able to see more completely the underlying aberrations and abnormalities that are impacting the tumor genome.

An organized listing of the HRD events is displayed within NxClinical software in a sample-info pop-up window.
Extract HRD Scores From Your Existing Data and Workflow
Another massive benefit of NxClinical 6.2 is it may be possible to extract these scores from the data and workflow you have right now.
Many sites currently running a capture-based NGS library prep today can leverage NxClinical 6.2 to calculate HRD scores from genomic scars without disrupting their existing data pipeline. NxClinical 6.2 is also compatible with major array platforms enabling the ability to implement an integrated genomic scar analysis from existing array service. This helps teams overcome a major hurdle that, until now, has limited HRD analysis for many labs.
Ready to Bring Powerful HRD Analysis and Scoring Capabilities to Your Lab?
Request a free personalized demo of NxClinical software and see its HRD analysis workflow in action. We will connect on an initial consultation to answer questions and dive a little deeper before demonstrating the latest version of NxClinical software—either with example data or your own.
1 Stover EH, Fuh K, Konstantinopoulos PA, Matulonis UA, Liu JF. Clinical assays for assessment of homologous recombination DNA repair deficiency. Gynecol Oncol. 2020;159(3): 887-898. doi:10.1016/j.ygyno.2020.09.029
2 Iliakis G, Murmann T, Soni A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombinationnrepair: Implications for the formation of chromosome translocations. Mutat Res Genet Toxicol Environ Mutagen. 2015 Nov;793:166-75. doi: 10.1016/j.mrgentox.2015.07.001. Epub 2015 Jul 4.
PMID: 26520387.
3 The Oncologist, Volume 27, Issue 3, March 2022, Pages 167–174
4 Wagener-Ryczek S, Merkelbach-Bruse S, Siemanowski J. Biomarkers for Homologous Recombination Deficiency in Cancer. J Pers Med. 2021;11(7):612. Published 2021 Jun 28. doi:10.3390/jpm11070612
5 Abkevich, et al., Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer, Br. J. Cancer 107 (10) (2012)1776–1782
6 Birkbak, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer discovery 2, no. 4 (2012): 366-375.
7 Popova T, Manie E, Rieunier G, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 2012;72:5454–62
For Research Use Only. Not for use in diagnostic procedures.
© Copyright 2022 Bionano Genomics, Inc. All rights reserved. All trademarks are the property of Bionano Genomics, Inc. or their respective owners.
*This software is for research use only. It is designed to assist clinicians and it is not intended as a primary diagnostic tool. It is each lab’s responsibility to use the software in accordance with internal policies as well as in compliance with applicable regulations.

