Blog
Notes on Analyzing Affymetrix OncoScan™ Data in Nexus Copy Number
Thermo Fisher’s OncoScan CNV Assay (formerly Affymetrix OncoScan™ array) is designed to get copy number as well as somatic mutation information out of cancer samples, especially solid tumor samples from Formalin-Fixed, Paraffin-Embedded (FFPE) tissues. Nexus Copy Number software has been the exclusive software solution for the analysis of OncoScan™ data due to its advanced cancer copy number algorithm, ease of use, and function to integrate the somatic mutation results. OncoScan™ customers can get the specialized software version (Nexus Express for OncoScan™) directly from Thermo Fisher free of charge, which is good for basic OncoScan™ data analysis, or they can get the more advanced full version of Nexus Copy Number software.
With OncoScan™ CNV Assay, the data file to be imported into Nexus is the .OSCHP file. Older versions of OncoScan™ data (e.g. V2) has the file name extension .CBZIP, which is also supported in Nexus. When loading the .OSCHP files, there are two options for the users: Affymetrix OSCHP-SNP-FASST2 and Affymetrix OSCHP-TuScan, the former being the default algorithm and the latter being the Thermo Fisher (Affymetrix) implementation of the ASCAT algorithm. Affymetrix OSCHP-SNP-FASST2 algorithm allows users to adjust the copy number calling thresholds and reset diploid baseline if necessary, while Affymetrix OSCHP-TuScan applies the ASCAT algorithm and users can’t modify the setting parameters. For details on adjusting settings and resetting diploid baseline, refer to blogs Change Analysis Settings in Nexus and Diploid Recentering: When, Why and How.
A variety of useful QC metrics and statistics are displayed in the Data Set table after analysis of the data. These include the Quality score, which measures the noise level of the data, %aberrant cells and overall ploidy, as well as copy number events and %LOH for each sample.
The strength of Nexus has always been its capacity of analyzing large sample size, i.e. hundreds or thousands of samples can be loaded into Nexus to identify the common aberrations or differences between different sample types (e.g. tumor grade 1 vs 2). Here’s an example of the genome view of 8 samples (4 tumors and 4 normals). Besides the copy number events (blue for gain and red for loss) and allelic events (yellow for LOH and purple for allelic imbalance), somatic mutations are displayed as diamonds (different colors for different types of point mutation, such as silent, nonsense, missense). Due to the limitation of Nexus Express for OncoScan™, it lists the somatic mutations in the events table in the single sample window, but doesn’t display them in the whole sample view, as shown in this example.
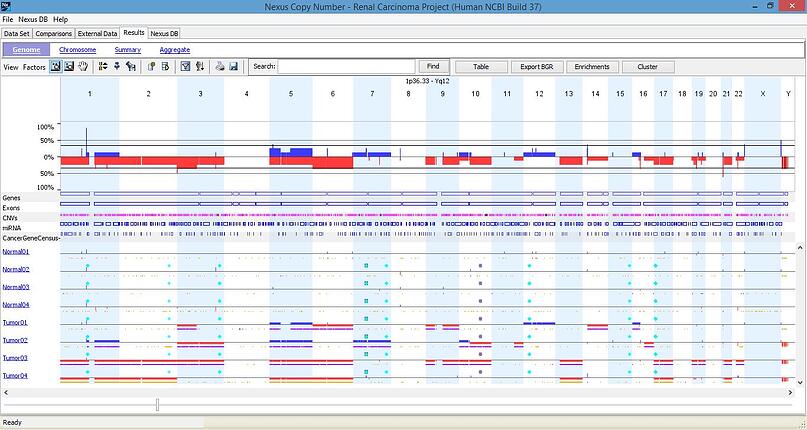
For the advanced downstream analyses, Nexus Express for OncoScan™ has the Comparison module that is for comparing different sample groups, while the full version of Nexus Copy Number has many more modules, including GISTIC, Survival analysis, Concordance function, Clustering, Enrichment analysis, etc. These modules provide the important tools for cancer researchers to go deeper with the copy number results. Please refer to other blog posts on the functions of these modules.
