Homologous Recombination Deficiency (HRD)
Integrate highly sensitive measurements of genomic instability associated with homologous recombination deficiency with Bionano solutions
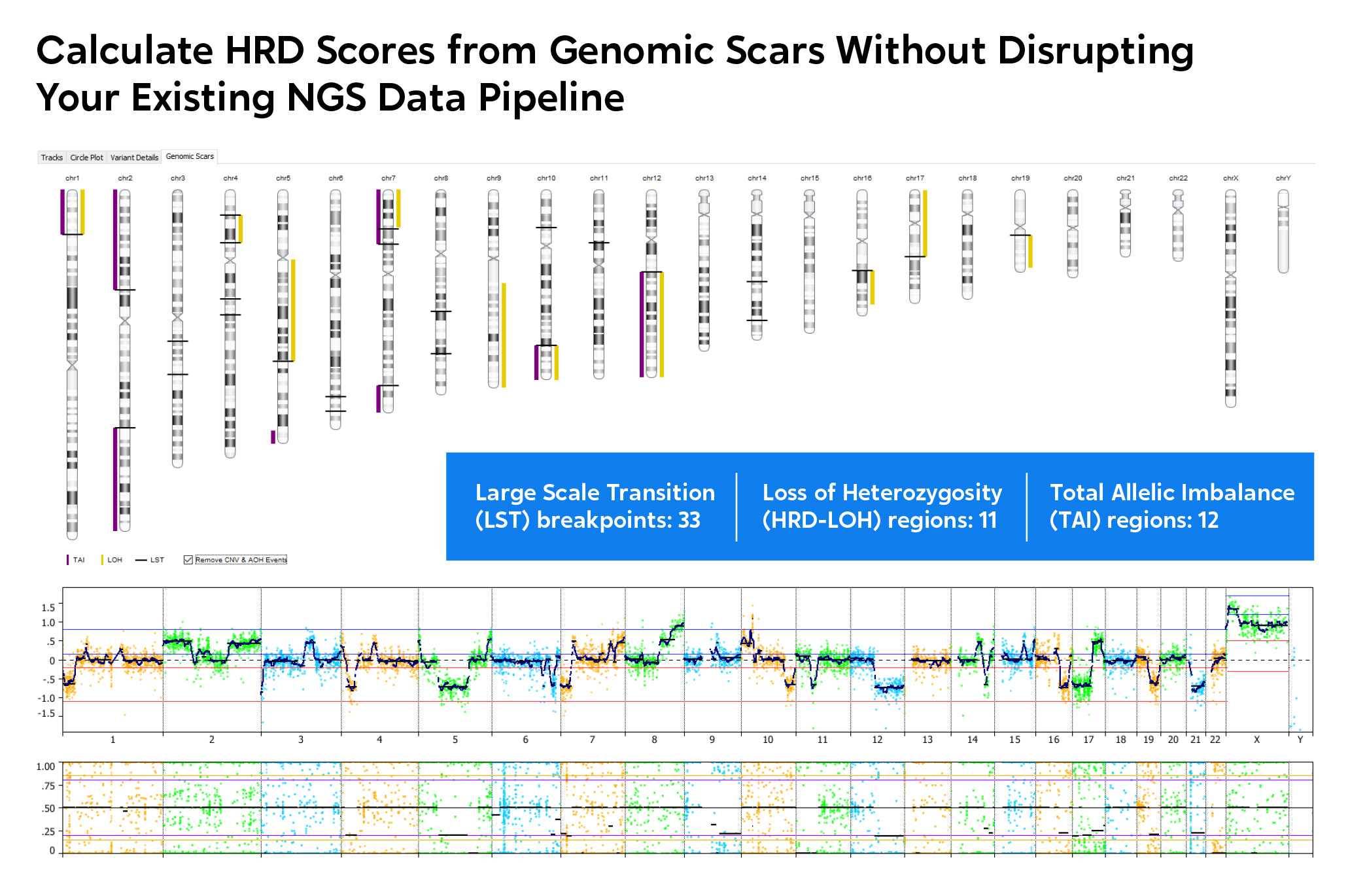
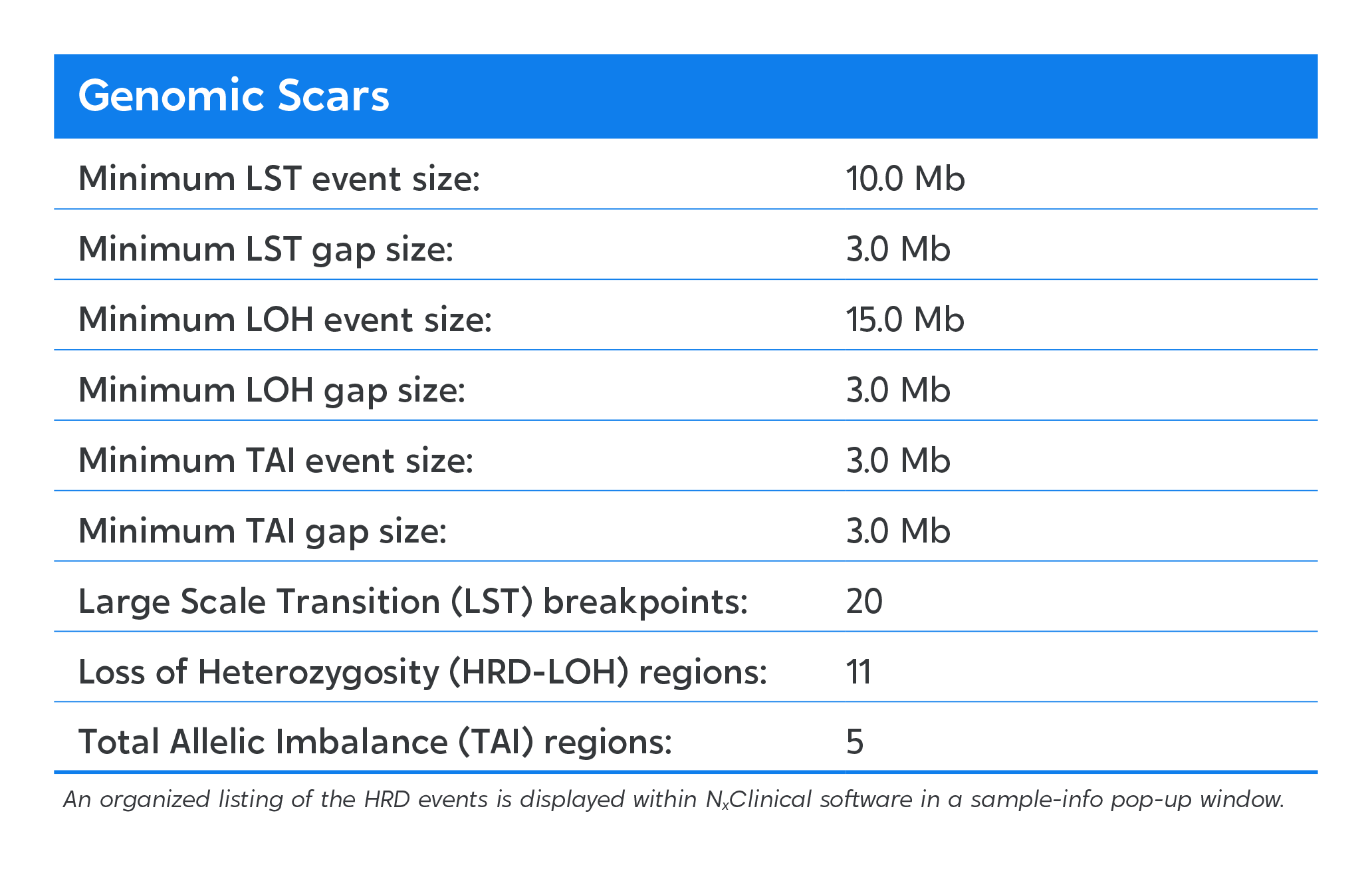
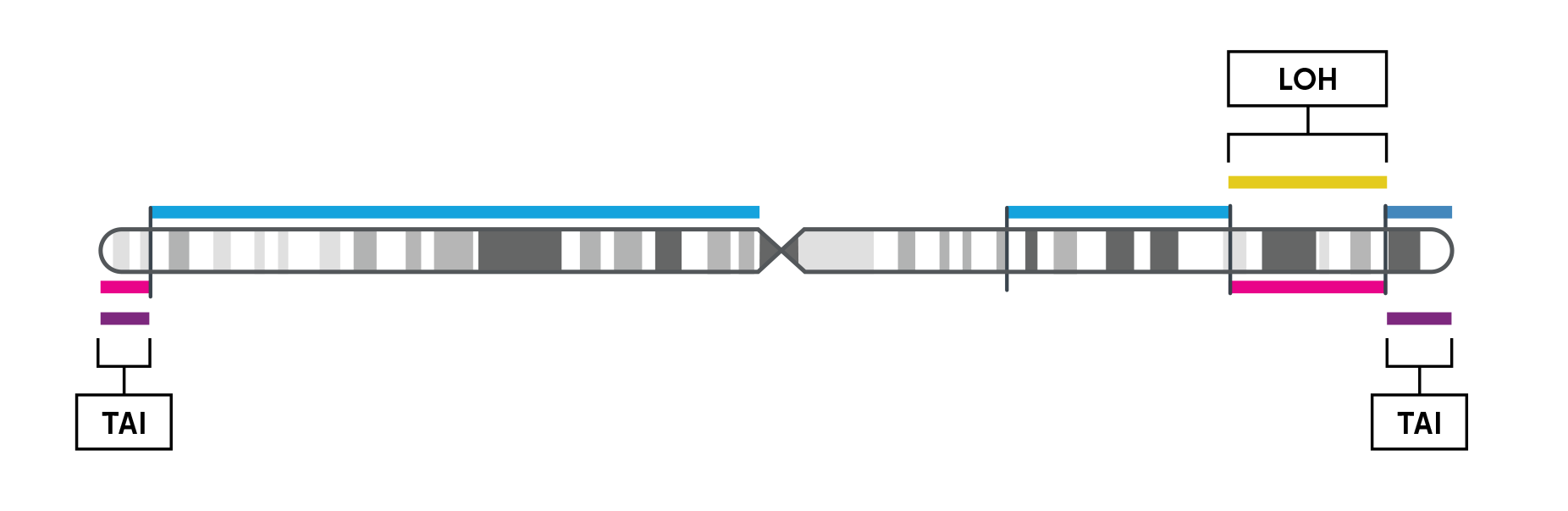
Homologous recombination deficiency (HRD) is a cell’s inability to repair double stranded DNA breaks resulting in an acquired rate of chromosome aberrations. HRD scoring is an analytical approach to measuring the cellular rate of acquiring chromosome breaks in specific, quantifiable patterns, or “genomic scars.” The three characteristic genomic scars are:
Cells with elevated HRD scores have been shown to be more sensitive to poly (ADP-ribose) polymerase (PARP) inhibitors and platinum-based therapies.
Whether you are looking to assess HRD profiles in hematological malignancies or solid tumors, Bionano offers different solutions that can help you achieve high-resolution and accurate analysis of genomic scar patterns, for accurate HRD assessment and stratification of samples. Leverage Bionano’s Saphyr® OGM workflow to assess all classes of structural variants in tumor samples and generate in-depth HRD assessments. Achieve detailed data visualization and HRD scoring, from multiple data types, such as sequencing and microarray data, with our VIA™ software for variant calling, annotation, and interpretation.
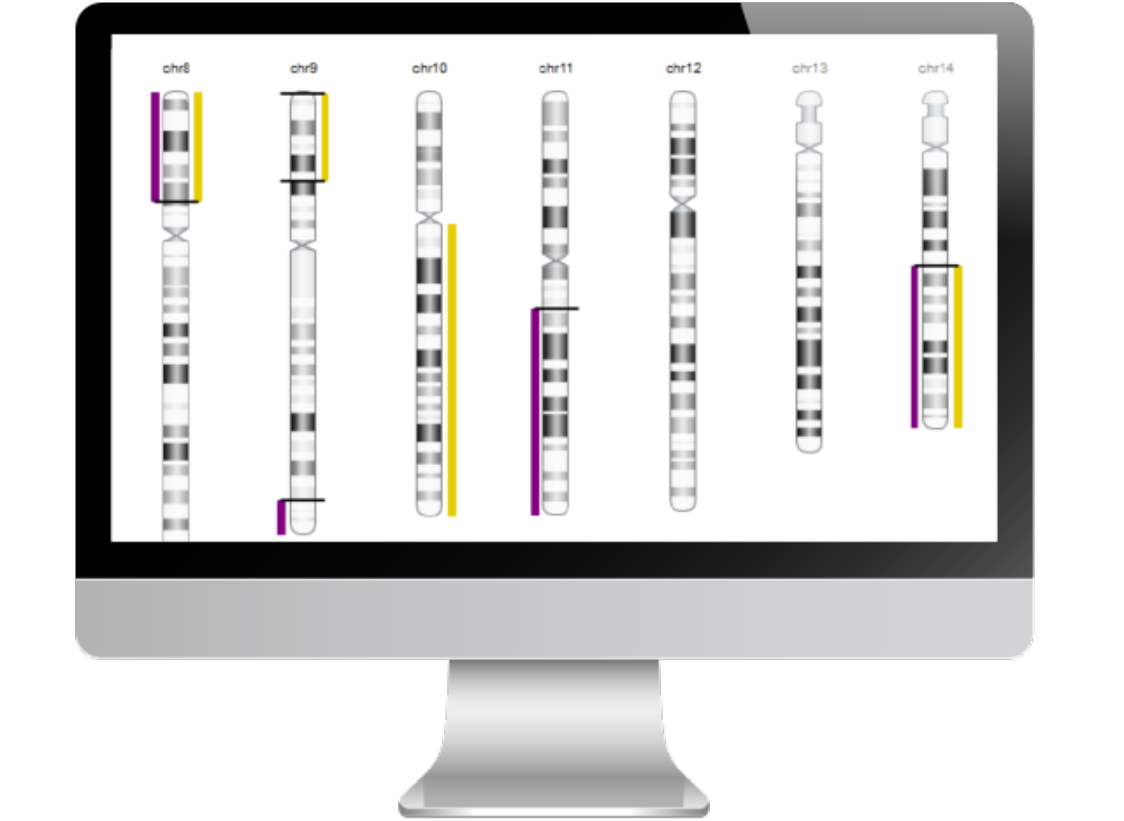


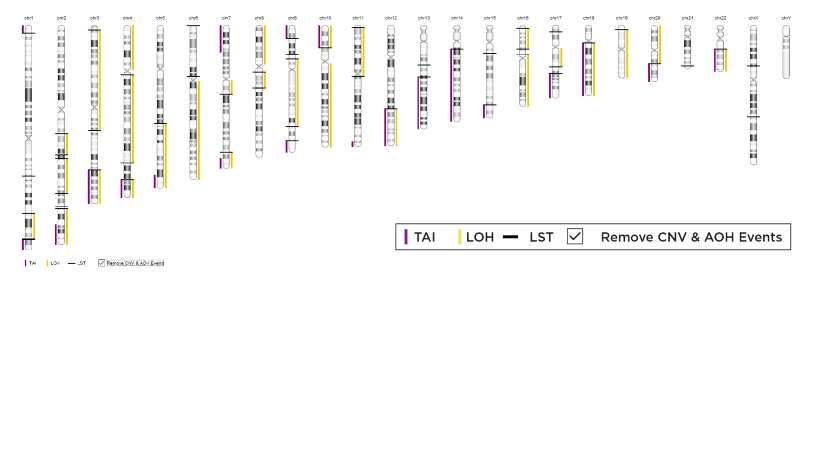
Chromosome-level visualization of genomic scars.
Genome-wide visualization of genomic scars.
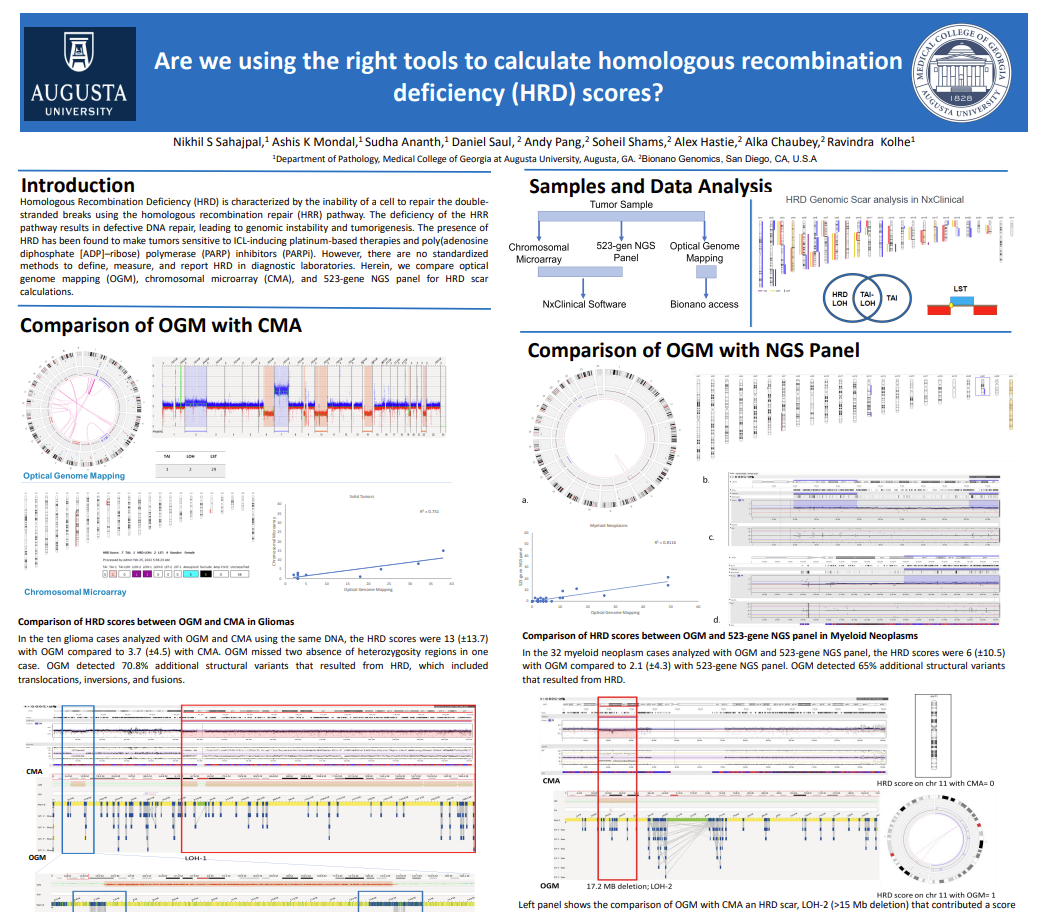
Data presented in poster format during the Association of Molecular Pathology meeting, 2022.
Learn more about an approach to genomic discovery that processes NGS data via VIA software to assess solid tumors for genomic instability states associated with HRD.
Watch WebinarSign up to watch the AMP 2022 Bionano Corporate Workshop #2 about the Comprehensive Assessment of HRD from Next-Generation Sequencing and Optical Genome Mapping.
Watch Workshop Recording
